The research conducted by the members of the diverse research groups in the Department of Biological Science is introduced here. The Department of Biological Science consists of 16 research groups, originating from the Biology Department founded in 1942 and the Molecular Biology Experimental Facility established in 1961. In addition, the department collaborates with 10 additional research groups affiliated with the Faculty of Science’s Sugashima Marine Biological Laboratory, the Center for Gene Research, the Institute of Transformative Bio-Molecules (ITbM), and the Graduate School of Pharmaceutical Sciences, making it possible to undertake cutting-edge interdisciplinary research and to provide a broadly-based undergraduate and graduate education.
Research Groups
- Department of Biological Science
- Sugashima Marine Biological Laboratory
- Nagoya University Center for Gene Research
- Institute of Transformative Bio-Molecules (ITbM)
- G30 International Program
- Graduate School of Pharmaceutical Sciences
*When sending email, please replace (at) with @.
Department of Biological Science
Group of Intercellular Signaling Biology
Plant survival strategies probed by identification of novel hormones
Hormones are molecules that are produced in the body and regulate various biological functions at very low concentrations, and their importance has long attracted the interest of researchers in both plants and animals. Using a combination of genomic and biochemical analyses, we have discovered five novel peptide hormones and their receptors that are involved in plant growth and environmental adaptation. We have also found that plants have a unique long-range signaling mechanism mediated by peptides that translocate in the phloem. Our research is also focused on elucidating the signaling pathways downstream of the receptors. By studying the function of each hormone and its signaling pathway, our goal is to decipher the unique and sophisticated survival strategies of sessile plants.
The expanding world of peptide hormones
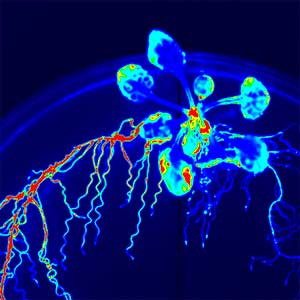
Over 25 years of research, we have discovered five novel groups of secreted peptide hormones involved in plant growth and environmental responses, including RGF, which is involved in root development, CEP, which communicates root nitrogen deficiency to the aboveground, and PSY, which regulates growth and stress response switching at the cellular level (Figure 1). We have also discovered the existence of long-distance-mobile non-secreted peptides such as CEPDL2, which communicates leaf nitrogen deficiency to the roots via phloem (Fig. 2, left). Although only a few plant hormones are listed in high school textbooks, the number of peptide hormones found in the past 25 years exceeds 20. By examining each peptide hormone in detail, we are amazed at how sophisticatedly plants engage in hormone- and receptor-mediated intercellular signaling during growth and environmental adaptation.
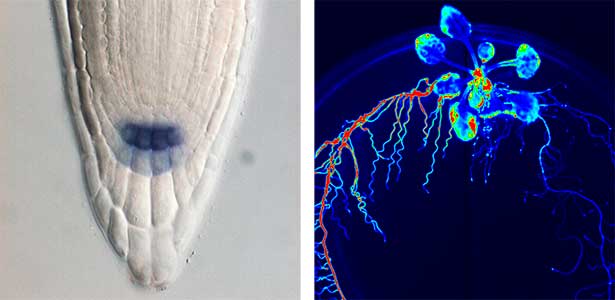
Fig. 1. Expression pattern of peptide hormone RGF required for root development (left). Systemic nitrogen demand signaling mediated by the peptide hormone CEP (right). Nitrogen deprivation of the right root causes a complementary upregulation of nitrate transporter expression in the left root. The peptide hormone CEP, which communicates nitrogen deficiency from roots to leaves, is involved in this process.
Post-translational modification enzymes and protein phosphatases
Various post-translational modifications are observed in extracellular secreted peptides and proteins, including peptide hormones, such as sulfation of tyrosine and hydroxylation of proline, as well as arabinosylation and galactosylation of hydroxyproline. All of these modifications are necessary to change the structure of peptides and proteins to achieve specific functions. We have succeeded in identifying the enzymes involved in these modifications and are investigating the physiological significance of post-translational modifications by observing the phenotypes of mutant plants deficient in these enzymes in detail (Fig. 2, right). In addition, protein phosphatases often play important roles downstream of peptide hormone receptors. Based on the identification of the substrates of protein phosphatases, we aim to discover new signaling pathways mediated by protein dephosphorylation.
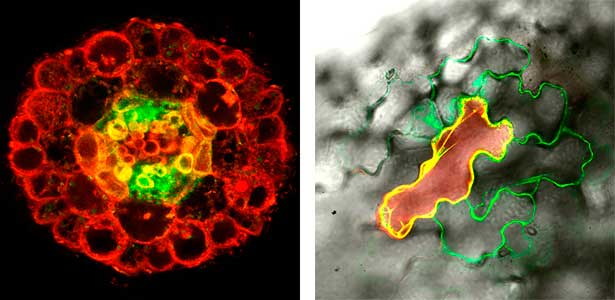
Fig. 2. Peptide CEPDL2 communicates aboveground nitrogen deficiency to the roots via phloem (left). The green GFP-CEPDL2 signal is seen in the phloem in a transverse section of a root. Physiological function of galactosylation of hydroxyproline (right). In a mutant plant deficient in galactosyltransferase, the permeability of the plasmodesmata is increased and molecular movement between cells is facilitated.
References
1. Ogawa-Ohnishi M. et al. Science 378, 175 (2022)
2. Ohkubo Y. et al. Nature Plants 7, 310 (2021)
3. Ohkubo Y. et al. Nature Plants 3, 17029 (2017)
4. Nakayama T., Shinohara H. et al. Science 355, 284 (2017)
5. Tabata R. et al. Science 346, 343 (2014)
6. Matsuzaki Y. et al. Science 329, 1065 (2010)
7. Ogawa M. et al. Science 319, 294 (2008)
8. Matsubayashi Y. et al. Science 296, 1470 (2002)
Member
-

Professor
Yoshikatsu Matsubayashi
matsu(at)bio.nagoya-u.ac.jp
Peptide signaling-mediated mechanisms of plant morphogenesis and environmental adaptation
-

Assistant Professor
Mari Ogawa-Ohnishi
ohnishi(at)bio.nagoya-u.ac.jp
Functional analysis of intercellular signaling focusing on post-translational modification enzymes
-

Assistant Professor
Yuri Ohkubo
ookubo.yuuri.n6(at)f.mail.nagoya-u.ac.jp
Environmental adaptation mechanisms in plants mediated by protein phosphorylation and dephosphorylation
Group of Plant Cell Biology
Plant cell morphogenesis
Plants are made up of cells surrounded by rigid cellulosic cell walls. Properly patterned cell walls are the determinant of plant cell shape, being the basis for the development of tissues and organs of plants. The cytoskeleton plays a central role in guiding the deposition of cell walls. Our research aims to understand the fundamental mechanisms underlying cell shaping, including molecular signaling pathways that orchestrate the assembly of cytoskeletal structures and their dynamic behaviors.
Patterning of cell walls in xylem vessel elements
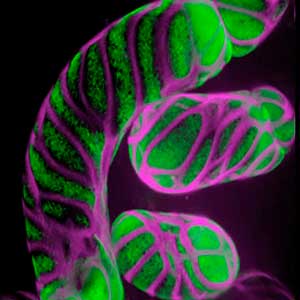
Differentiating ylem vessel elements deposit thick, hydrophobic secondary cell walls and undergo programmed cell death, forming a hollow tubular structure. Prior to the secondary cell wall deposition, cortical cytoskeletons are rearranged into distinct patterns to direct the deposition of the cell walls in the annular, spiral, reticulate, and pitted patterns. We study the molecular signaling that governs the dynamic behavior of cytoskeletons in differentiating xylem vessel elements. (Fig. 1).
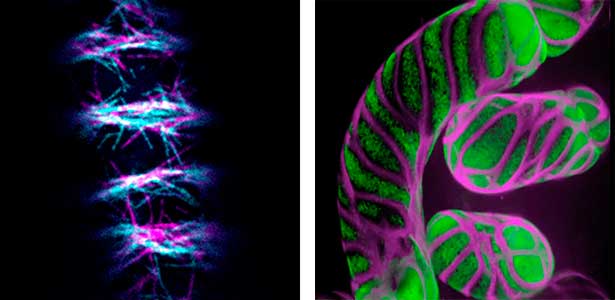
Fig. 1. Microtubules (left) and cell walls (right, magenta) in differentiating xylem vessel elements.
Plant cytokinesis
Plant cells undergo cytokinesis by forming a cell plate. The position and direction of the cell plate formation determine the shape and subsequent cell fate of the daughter cells. The plant-specific cytoskeletal structures, such as prophase microtubule bundles (PPBs) and phragmoplasts, direct cell plate formation (Figure 2). We study the mechanisms underlying the assembly and action of the PPB and phragmoplasts and their evolutionary aspects.
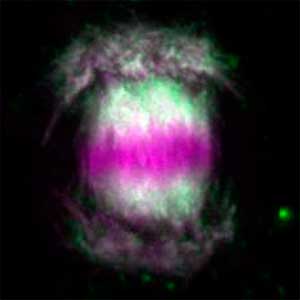
Fig. 2. Phragmoplast microtubules (magenta) and microtubule-associated proteins (green).
References
1. Kijima ST. et al. Nature Commun. 16, 1921 (2025)
2. Higa T. et al. Nature Plants 10, 100 (2024)
3. Sasaki T. et al. Nature Commun. 14, 6987 (2023)
4. Sasaki T. et al. Curr. Biol. 29, 4060 (2019)
5. Sugiyama Y., Nagashima Y. et al. Nature Commun. 10, 468 (2019)
6. Oda Y. and Fukuda H. Science 337, 1333 (2012)
Member
-

Professor
Yoshihisa Oda
oda.yoshihisa.w5(at)f.mail.nagoya-u.ac.jp
Plant cell morphogenesis
-

Assistant Professor
Takema Sasaki
sasaki.takema.w0(at)f.mail.nagoya-u.ac.jp
Cytokinesis in land plants
-

YLC Designated Assistant Professor
Yuki Sugiyama
sugiyama.yuki.d8(at)f.mail.nagoya-u.ac.jp
Autolysis of phloem sieve elements
Group of Reproductive Biology
Understanding of the mechanisms regarding sex and reproduction
Organisms employ many types of sex determination: Genetic sex determination and environmental sex determination. Organisms that changes sex are not rare and organisms that produce offsprings without male are present in nature. Why do most organisms have sex? How have most organisms been creating such a variety of reproductive systems - this is one of the largest mysteries in biology. Our lab has been revealing the molecular and cellular mechanisms of how a single fertilized egg develop into female or male and how germ cells fit the sex of body to produce eggs or sperm. Our investigation has been clarifying the meaning of sex and the evolutional bases that create a variety of reproduction systems.
Core mechanism of sex determination
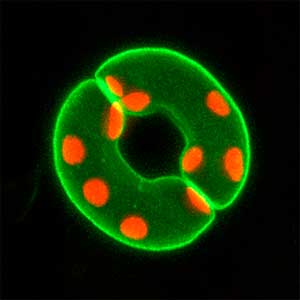
We have been revealing that sex is determined in the balancing of feminization and masculinization. Once the sex looks determined, the body still retains the potential to develop into female or male and the tug of war between the two sexes continue. This principle leads to sex changes (reversal). We have found that germ cells are critical for feminization and in the absence of the germ cells the somatic cells are predisposed to male development.
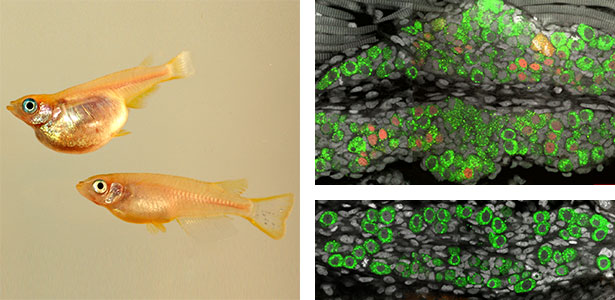
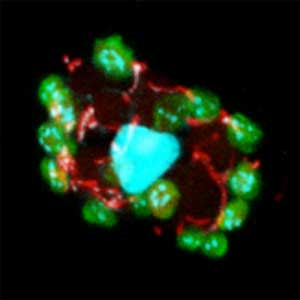
Fig. 1. Sex reversed female medaka from male with an excess amount of germ cells (upper left) and normal male (lower left). More germ cells (green) are present in female gonads (upper right) than in male testes (lower right).
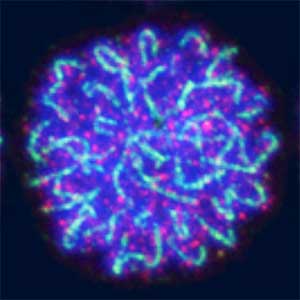
Sex determination of germ cells towards eggs or sperm
Sex determination triggers modules in germline stem cells that promote eggs or sperm development. Eggs and sperm are continuously produced from sexually indifferent germline stem cells in ovary and testis. Then how do germline stem cells determine the sexual fate of descendant germ cells? We have identified the sexual switch gene in germ cells, foxl2l (foxl3). We have also revealed that foxl2l initiates several modules that correspond to female-specific characters. During the analyses, we have found the evidence suggesting evolutional driving force may be different between eggs and sperm.
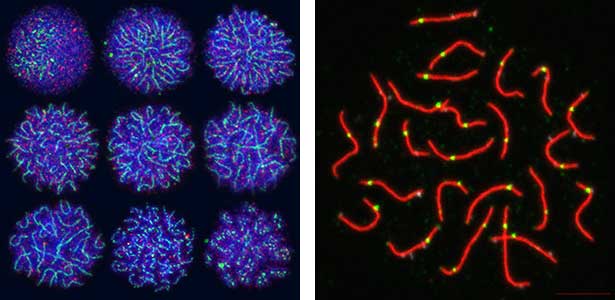
Fig. 2. Medaka chromosomes at various stages of meiosis (left) and after homologous recombination (right; green dots indicate crossover sites).
Molecular basis that allows a variety of reproduction - How parthenogenic species appear during evolution
Several modules functioning during oogenesis and spermatogenesis are genetically independent, which allows combination of different modules to function in germ cells. This raises hypothesis that combination leads to different types of reproduction systems. Our lab is addressing a critical event that creates parthenogenetic species by investigating the change of modules using medaka and Poecillia. We are investigating a variety of reproduction in terms of change of modules.
References
1. Kikuchi M. et al. Development 151, dev201840 (2024)
2. Kikuchi M. et al. PNAS 117, 12174 (2020)
3. Nishimura T. et al. PLoS Genet. 14, e1007259 (2018)
4. Nishimura T. et al. Science 349, 328 (2015)
5. Nakamura S. et al. Science 328, 1561 (2010)
6. Kurokawa H. et al. PNAS 104, 16958 (2007)
Member
-

Professor
Minoru Tanaka
tanaka.minoru.r3(at)f.mail.nagoya-u.ac.jp
Molecular mechanisms of sex determination and differentiation
-

Assistant Professor
Mariko Kikuchi
kikuchi.mariko.g0(at)f.mail.nagoya-u.ac.jp
Molecular mechanisms underlying sexual and reproductive diversity
-

GSS Designated Assistant Professor
José Carranza Luna
carranza.jose.g9(at)f.mail.nagoya-u.ac.jp
Molecular mechanisms of germ cell masculinization and cortisol on reproduction
Group of Plant Physiology
Elucidation of the environmental response in plants
Since plants cannot move, they must accurately respond to the changing environmental conditions around them, such as light, water and nutrient levels of soil, carbon dioxide (CO2), temperature, etc., in order to grow. Stomata in the plant epidermis play a crucial role by opening and closing in response to various environmental signals, regulating gas exchange between plants and the atmosphere, including the uptake of CO2 for photosynthesis, transpiration, and the O2 release. Stomata open in response to sunlight, especially blue light acting as a signal and red light inducing photosynthesis, to enhance CO2 uptake. When the plants are exposed to drought stress, stomata close in response to the plant hormone abscisic acid to prevent water loss from the plants. In addition, stomata are known to close in response to elevated CO2 levels. We aim to advance research using stomata as a model material for environmental response and unravel the signal transduction mechanisms in plant environmental responses.
Light signal transduction and control of plant growth
We have pioneered the discovery that the plant-specific blue light receptor, phototropin, serves as a light receptor for stomatal opening. The blue light signal received by phototropin induces the activation of the plasma membrane H+-ATPase, leading to the formation of the driving force for stomatal opening. (Fig. 1). Currently, we are actively conducting physiological, biochemical, and genetic analyses, as well as synthetic biology research using guard cells around stomata, aiming for a comprehensive understanding of the signaling mechanism controlling stomatal aperture. As member of the Institute of Transformative Bio-Molecules (ITbM), we also explore research from a chemical biology perspective. Moreover, we are advancing research on stress-resistant plants and increased crop yield through the manipulation of stomatal aperture based on the results from basic research.
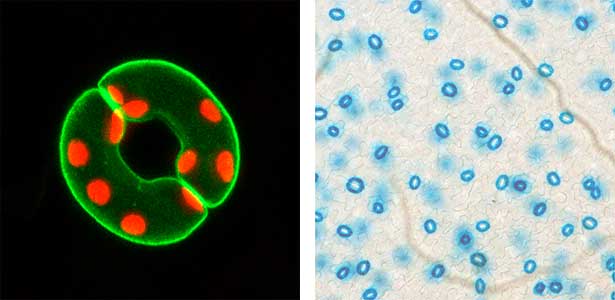
Fig. 1. Localization of plasma membrane H+-ATPase, a key enzyme for stomatal opening, in guard cells (Left). Green fluorescence represents GFP-H+-ATPase and red fluorescence represents chloroplasts. GUS-staining of stomatal guard cells using guard cell-specific promoter (Right).
Water stress- and CO2-induced responses in plants
Land plants prevent excessive water loss through transpiration by closing stomata. Stomatal closure in response to an increase in CO2 concentration is crucial for improving plant water use efficiency. The CO2 sensor complex within stomatal guard cells detects this increase (Fig. 2) and then initiates intracellular signal transduction, leading to stomatal closure. Our goal is to understand the function and molecular mechanisms of plant CO2 sensing and signal transduction. We’re employing biochemical, genetic, and cell biological approaches, collaborating with experts in structural biology, chemical biology, mass spectrometry, and genome evolution to explore the principles of cellular function.
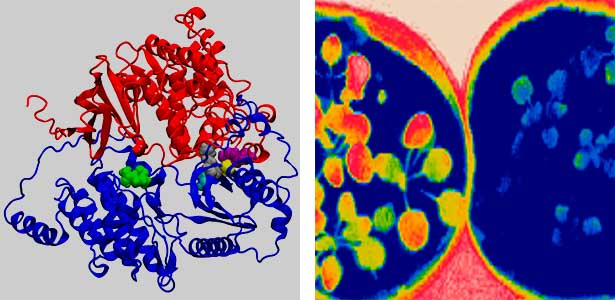
Fig. 2. Three-dimensional structural model of the Arabidopsis CO2 sensor (left) and thermographic images of Arabidopsis plants (right). The CO2-insensitive mutant (right pot) does not show stomatal closure even at high CO2 concentrations and shows a significant decrease in leaf temperature due to transpiration compared to the wild type (left pot).
References
1. Hayashi Y. et al. Nature Commun. 15, 1194 (2024)
2. Aihara Y. et al. Nature Commun. 14, 2665 (2023)
3. Takahashi Y. et al. Science Adv. 8, eabq6161 (2022)
4. Zhang M. et al. Nature Commun. 12, 735 (2021)
5. Kinoshita T. et al. Nature 433, 167 (2005)
6. Kinoshita T. et al. Nature 414, 656 (2001)
Member
-

Professor
Toshinori Kinoshita
kinoshita(at)bio.nagoya-u.ac.jp
Signaling pathways in response to environmental signals in plants
-

Designated Associate Professor
Yohei Takahashi
ytakahashi(at)itbm.nagoya-u.ac.jp
Water stress- and CO2-induced responses in plants
-

Assistant Professor
Koji Takahashi
takahashi(at)bio.nagoya-u.ac.jp
Signaling pathways in cell elongation in plants
-

Assistant Professor
Yuki Hayashi
y-hayashi(at)bio.nagoya-u.ac.jp
Analysis of environmental response mechanisms in plants using guard cells as a model
-

Assistant Professor
Eigo Ando
ando.eigo.j9(at)f.mail.nagoya-u.ac.jp
Mesophyll-to-guard cell signal transduction in response to photosynthesis
Group of Neural Circuit
Understanding the neural mechanisms controlling animal behaviors
Mate selection behaviors are universally observed in the animal kingdom, with many animals using sensory communication during the selection process. Our research focuses on uncovering how specific sensory stimuli are processed in the brain to extract 'meaning', enabling successful communication with mates. We're also delving into the mysteries of how sensory processing, responsible for this communication, has evolved across species and how it contributes to inter-species differentiation. This is a collaborative endeavor within our research group. We use both 'Drosophila' (fruit flies) and mosquitoes for our experiments —species with simple brains that are easy to observe and experiment with.
Neural mechanisms underlying auditory communication
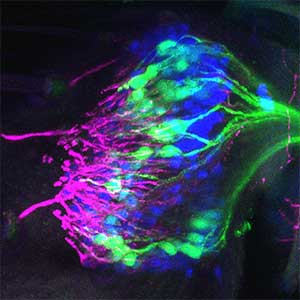
As part of courtship behavior, many animals engage in communication using sound. In this context, the receiver animal evaluates species-specific acoustic signals, often termed ‘courtship songs’, to determine whether to accept the sender as a potential mate. It remains unclear how the brain recognizes and processes these distinct courtship song signals emitted by conspecifics. We aim to address this topic by elucidating the properties of neurons responsible for processing auditory information and unraveling the associated neural circuitry (Fig. 1). Our research, employing molecular genetics in studies using Drosophila, aims to comprehend the neural mechanisms that evaluate sound. Additionally, by applying these findings to mosquitoes, we aim to control populations of various disease-transmitting mosquitoes, contributing to disease prevention.
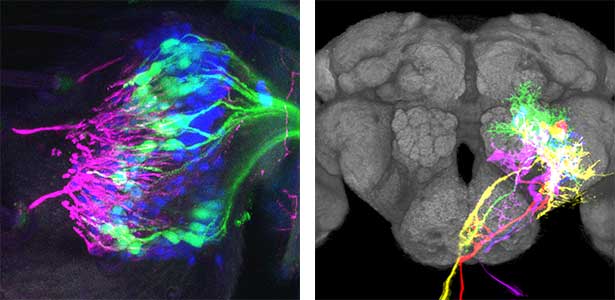
Fig. 1. Auditory sensory neurons in the fly ear (left) and neurons responsible for processing auditory information in the fly brain (right).
Neural mechanisms behind the evolution of animal behavior
Animal behaviors, and the signals used in communication, have diversified across species. How has the brain of animals evolved to efficiently perceive and extract signals from conspecifics? We focus on the diversification of signals such as sound and pheromones used in Drosophila courtship behavior across different species. By comparing the neurons and neural circuits of species utilizing different signals, we aim to unravel this mystery. Furthermore, our recent investigations involve studying how insect brains, particularly those of Drosophila breeding within flowers, have evolved to perceive signals from flowers (Fig. 2).

Fig. 2. Courtship behaviors of Drosophila elegans that breed within flowers.
Neural mechanisms controlling instinctive behaviors in insects
Despite having simple nervous systems, insects thrive and reproduce in diverse environments across the globe. To understand how insects have diversified their instinctive behaviors to ensure successful reproduction and survival, we study three important behaviors within the Drosophilidae family: courtship, mating, and social aggregation. We seek to uncover the neural mechanisms behind these behaviors, exploring how they have evolved and diversified, with diversification of instinctive behaviors evident even among Drosophila species. By comparing genes and neural circuits between the model species Drosophila melanogaster and other Drosophila family members, we aim to shed light on the mechanisms driving the evolution of instinctive behaviors.
References
1. Tanaka R. et al. Commun. Biol. 7, 1714 (2024)
2. Imoto K. et al. iScience 27, 110266 (2024)
3. Loh Y.M., Xu Y.Y.J. et al. iScience 27, 110264 (2024)
4. Yamanouchi M.H. et al. iScience 26, 106617 (2023)
5. Ohashi S.T. et al. Sci. Rep. 13, 383 (2023)
Member
-

Professor
Azusa Kamikouchi
kamikouchi.azusa.r4(at)f.mail.nagoya-u.ac.jp
Neural circuit controlling auditory communication behaviour
-

Lecturer
Yuki Ishikawa
ishikawa.yuki.e2(at)f.mail.nagoya-u.ac.jp
Neural basis of animal behavioural evolution
-

Lecturer
Ryoya Tanaka
tanaka.ryoya.z3(at)f.mail.nagoya-u.ac.jp
Neural mechanisms controlling instinctive behaviour in insects
-

YLC Designated Assistant Professor
Matthew P. Su
su.matthew.paul.y3(at)f.mail.nagoya-u.ac.jp
Neural mechanisms controlling auditory behaviour in mosquitoes
Group of Organ Function
Studies of developmental biology and neuroscience using zebrafish and medaka
The developmental process of vertebrate embryos, which involves the formation of organs with complex structures and functions from fertilized eggs, is precisely controlled. Neural tissue is induced from the dorsal ectoderm. In this tissue, individual neural regions are determined along the anterior-posterior axis, and neural stem cells and neural precursor cells are generated in those regions, leading to the differentiation of neurons. These neurons migrate, extend their neurites, and form neural circuits. Additionally, on the dorsal side of the neural tissue, there are stem cells known as neural crest cells, which differentiate into various cell types, including pigment cells. In our laboratory, we use small fish species such as zebrafish and medaka to investigate the mechanisms of neuronal and pigment cell differentiation, as well as the functions of neural circuits.
Neurogenesis-cerebellum development
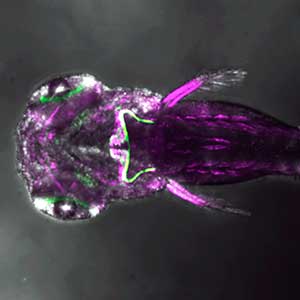
The structure of the cerebellum and cerebellar neural circuits in vertebrates is relatively conserved from fish to mammals. During development, the major neurons of the cerebellum, granule cells and Purkinje cells (Fig. 1. left panel), are generated from neural precursor cells located in the dorsal and ventral parts of the cerebellar primordium, respectively. Granule cells and Purkinje cells receive two different types of input fibers from outside the cerebellum (mossy fibers and climbing fibers). These two types of information are integrated in the Purkinje cells and eventually output to the outside of the cerebellum. We believe that this simple scheme is a good model for understanding the mechanisms of neural circuit formation in the brain. We are using transgenic zebrafish to visualize cerebellar neural circuits (Fig. 1. right panel) and creating mutants using genome editing techniques to elucidate the molecular mechanisms of differentiation of cerebellar neurons and neurons that input to the cerebellum, as well as the formation of cerebellar neural circuits.
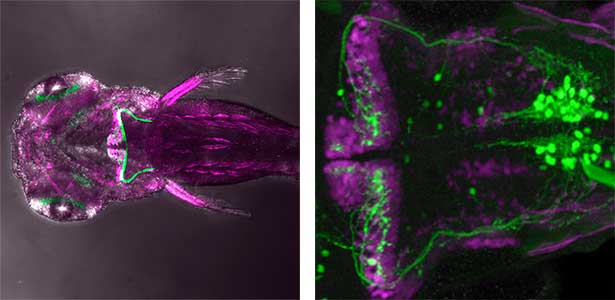
Fig. 1. Major neurons in the zebrafish cerebellum (left). Green represents the axons of granule cells and magenta represents Purkinje cells. Visualization of cerebellar neural circuits (right). Green represents climbing fibers, and magenta represents Purkinje cells.
Neural circuit function-fear response learning, social behavior
The cerebellum has been thought to be involved in coordinated motor control and motor learning. Recent studies have revealed that the cerebellum is also involved in higher brain functions such as emotions like anxiety and fear, as well as cognition. Furthermore, abnormalities in cerebellar neural circuits are thought to be related to human mental disorders such as autism spectrum disorder (ASD). We are using zebrafish to analyze the role of cerebellar neural circuits in motor learning, fear response learning, and social behavior. We aim to clarify the higher functions of cerebellar neural circuits by performing functional imaging of these circuits and manipulating their activity by expressing neurotoxins or optogenetic tools in cerebellar neural circuits.
Neural crest cell differentiation-pigment cell differentiation
Pigment cells are specialized cells in vertebrates that create body color and patterns. In mammals and birds, pigment cells consist of only one type, melanocytes, which produce melanin. However, other vertebrates have melanophores (pigment cells that exhibit black color) homologous to melanocytes, as well as iridophores and xanthophores. Additionally, some teleost fish have a variety of pigment cells, including leucophores and cyanophores. These pigment cells, like melanocytes, differentiate from a common pigment stem cell derived from neural crest cells. Therefore, teleost fish have more types of pigment cells differentiating from pigment stem cells compared to mammals and birds. We are using fish, particularly medaka, as a model for pigment cells, and using mutants (Fig. 2) to elucidate the mechanism by which a variety of cell types are produced from stem cells.

Fig. 2. Medaka with wild-type body color (left) and mutant medaka that do not develop melanophores, xanthophores, or leucophores (right).
References
1. Itoh T. et al. Development 151, dev202546 (2024)
2. Hagio H., Koyama W. et al. eLife 12, e83975 (2023)
3. Hagio H., Koyama W. et al. eLife 12, e83974 (2023)
4. Miyadai M. et al. Development 150, dev202114 (2023)
5. Koyama W. et al. eNeuro 8, ENEURO.0507-20.2021 (2021)
Member
-

Professor
Masahiko Hibi
hibi.masahiko.s7(at)f.mail.nagoya-u.ac.jp
Elucidation of the mechanisms of higher-order brain structure formation
-

Associate Professor
Takashi Shimizu
shimizu.takashi.k2(at)f.mail.nagoya-u.ac.jp
Elucidation of the functional mechanisms of cerebellar neural circuits
-

YLC Designated Assistant Professor
Takuya Kaneko
kaneko.takuya.n5(at)f.mail.nagoya-u.ac.jp
Wiring mechanism of vagal circuits for brain-body communication

Group of Integrative Ecology and Evolution
Evolution of sexual and social traits
Animals show amazing diversity in their behavior and morphology. Especially, sexual selection and social cooperation are the keys to understand the evolution of conspicuous ecological traits. Our goal is the integrative evolutionary understanding of these ecological characters in terms of genetic, developmental and physiological mechanisms. We mainly focus on insects, and investigate the ecological significance of behavior, genetic mechanisms underlying the trait expression, and molecular mechanism of phenotypic plasticity. Key techniques are RNAi-mediated gene knockdown, morphology, histology, behavioral observation, bioinformatics and field works.
Evolution of beetle weapons and its phenotypic plasticity
Large number of animals have evolved sexual weapons and ornaments via sexual selection. Beetle weapons (e.g. horns and mandibles) are one of the best examples of sexually selected exaggerated traits. These weapons are effective in male-male combat but costly in normal life. Therefore, large weapons only develop in well-nourished large males, and small males that have grown up in poor conditions develop tiny, rudimental weapons. This striking plasticity is a common characteristic of sexual weapons. By using broad-horned flour beetle (Gnatocerus cornutus), we clarified that histone deacetylases (HDACs) regulate the mandible size plasticity. As an internal physiological signal of nutritional state, a growth factor called insulin-like peptide 2 (ILP2) is shown to control the nutrition-dependent weapon growth in broad-horned flour beetle.

Fig. 1. Broad-horned flour beetle (Gnatocerus cornutus). RNAi-mediated gene knockdown (KD) of insulin-like peptide 2 (ILP2) generates rudimentary mandible in males. Left: control (wild type), Right: ILP2-KD phenotype.
Social caste evolution in insects
he colony of social insects such as ants consists of various castes. Queen is a specialized egg-layer, whereas intranidal workers perform nursing and extranidal workers are engaged in foraging and defense. These behavioral and physiological differences of castes are basically driven by developmental plasticity. We aim to clarify how caste differentiation occurs in terms of social interaction, gene expression and physiological state. In case of monomorphic ponerine ant Diacamma cf. indicum, insulin-signaling is up-regulated in queen to rapidly produce eggs. In contrast, intranidal workers has lowered metabolic state and they store energy inside their body (e.g. storage protein hexamerin). By focusing on social interaction, gene expression and physiological heterogeneity, we are trying to elucidate the evolutionary mechanism of social behavior.
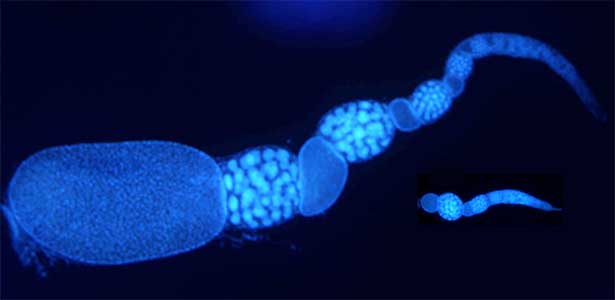
Fig. 2. Ovary of Diacamma ant. Socially dominant individuals develop ovarioles and become functional queen (left). In contrast, ovarian development is arrested in subordinate individuals (right).
References
1. Sugiyama, M. et al. PLoS Genetics 19.12: e1011069 (2023)
2. Okada, Y. et al. PLoS Biology 17(11): e3000541 (2019)
3. Ozawa,T. et al. PNAS 113, 15042-15047 (2016)
4. Okada, Y. et al. Molecular Ecology 26, 2922-2938 (2017)
5. Fujioka, H. et al. Biology Letters 13, 20160743 (2017)
6. Okada, Y. et al. Journal of Insect Physiology 56, 288-295 (2010)
Member
-

Professor
Yasukazu Okada
okayasukazu(at)gmail.com
Evolution of sexual and social traits

Group of Neuroethology
Neuronal principles of sleep and memory
We aim to elucidate the functions and mechanisms of sleep by investigating various animals, including reptiles. This neuroethological approach provides unique insights to identify general neuronal principles conserved across amniotes.
Are the brain circuits generating sleep conserved across species?
Humans cycle through Rapid Eye Movement (REM) sleep and non-REM (NREM) sleep approximately five times in 90-minute intervals during a night. Recently, it has been discovered that Australian dragons Pogona vitticeps also experience REM and NREM sleep. Remarkably, the cycle of REM/NREM sleep in dragons occurs roughly every 90 seconds, repeating over 400 times a night. While the cycle lengths vary greatly among animal species, the underlying “pattern generator” between REM and NREM sleep may be conserved across species. Revealing this entity is one of our major goals. We utilize various methods such as cutting-edge electrophysiological techniques, pharmacological methods, molecular biology techniques, and behavioral observations to advance our research.
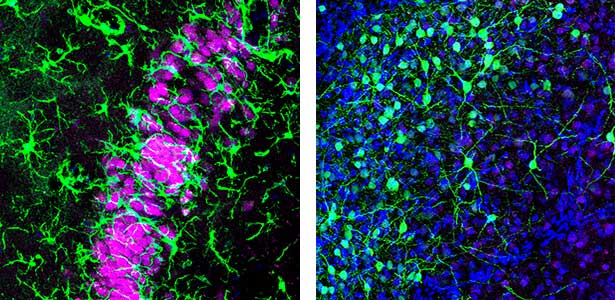
Fig. 1. Left, a lizard’s hippocampus exhibits a layered structure of neurons closely resembling that of the mammalian hippocampus. Right, neurons in the claustrum of a lizard. The claustrum is a mysterious brain region thought to be involved in consciousness. Recently, we have revealed that the claustrum is involved in generating neuronal activity during NREM sleep.
Mechanisms of memory consolidation during sleep
Our daily experiences are consolidated into memories in the brain during sleep. In recent years, it has been found that a brain oscillation called sharp wave ripple (SWR), generated in the hippocampus during sleep, “replay” neural activity that occurred during wakefulness, thereby consolidating memories. Furthermore, we have recently discovered that SWRs during sleep not only replay important memory information but also erase unnecessary information. This mechanism is believed to efficiently consolidate memories into the brain. Currently, we are diligently researching to further elucidate the function and dynamics of SWRs.
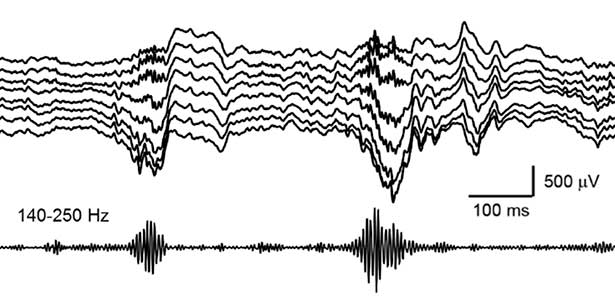
Fig. 2. Hippocampal sharp wave ripples (SWRs). The high frequency component called “ripple (140-250 Hz)” play a role in memory consolidation.
References
1. Norimoto H., Fenk L.A. et al. Nature 578, 413 (2020)
2. Norimoto H. et al. Science 359, 1524 (2018)
3. Shein-Idelson M. et al. Science 352, 590 (2016)
Member
-

Professor
Hiroaki Norimoto
norimoto.hiroaki.h3(at)f.mail.nagoya-u.ac.jp
Neuronal principles of sleep and memory
-

Assistant Professor
Yasutaka Mukai
mukai.yasutaka.z3(at)f.mail.nagoya-u.ac.jp
Neuromodulatory mechanisms underlying behavior generation
Group of Cell Regulation
Exploring human neuropsychiatric pathologies using mouse models
Actin, tubulin, and septin are polymerizing ATP/GTPases. In cooperation with motor proteins, these cytoskeletal components control cell shape and rigidity, and generate force for cell division/movement/morphological differentiation, etc. We focus on septins and related molecules that abound in the brain, and generate genetically engineered mice to explore unknown molecular mechanisms underlying behavioral abnormalities in sensorimotor, cognitive, and social functions. Our mice with specific abnormalities are useful as models of brain dysfunctions (e.g., amnesia, Parkinson's disease, and developmental disorders) to probe pathological states and develop diagnostic and therapeutic methods. Some of our ongoing research projects are shown below.
Exploring the interaction among genetic and environmental factors that influence human cognition by using mice and cultured neurons
A human genomic locus associated with our intelligence (IQ) contains SEPT3, a septin gene whose physiological function is unknown. Our systematic behavioral screening pinpointed premature memory decay in mice that lack Sept3. We discovered a unique abnormality in the hippocampal dentate gyrus (DG) synapses, which was recapitulated in cultured DG neurons. These mice and cultured neurons can be regarded as in vivo and in vitro models for mild cognitive impairment (MCI, or “forgetfulness,” the earliest symptom of dementia) in humans. Interestingly, the memory decay and morphological abnormalities of DG synapses in Sept3 knockout mice were ameliorated by rearing them in an enriched environment that promotes physical and social activities. Currently, we are searching for the metabolic and endocrine changes induced by environmental factors (exercise, dietary restriction) that are known to improve cognitive function including memory, and the mechanism by which their memory decay/synaptic abnormalities are improved.
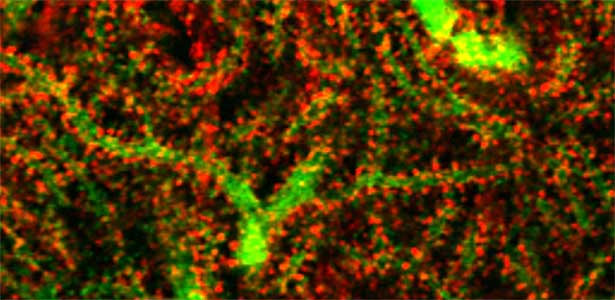
Fig. 1. Glial processes (red: CDC42EP4, a major binding partner of septin) enwrap synapses of Purkinje cell dendrites (green: calbindin) in the cerebellum. In mice that lack CDC42EP4, which serves as a scaffold for glutamate transporters, reuptake of glutamate is delayed and glutamate remains in the perisynaptic spaces, resulting in motor learning defects.
Modeling human intellectual disabilities in mice by recapitulating genetic mutations and analyzing the molecular mechanisms
Low molecular weight G proteins such as Rac/Rho/Cdc42 and their effectors regulate diverse cellular signaling pathways and cytoskeletal dynamics. The regulations are crucial for the brain development; ensuring progenitor cell division and migration, neurite protrusion and projection, and synaptogenesis. Countless genetic variations are found in pediatric patients with structural brain abnormalities, intellectual disabilities, and epilepsy, but only a few of them have been proved to be responsible for the pathology. Thus, we aim to screen pathogenic variants and elucidate the underlying molecular mechanisms by analyzing the brain structure and function in mosaic transgenic mouse fetuses/newborns.
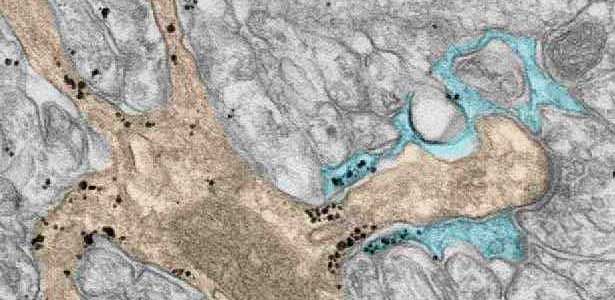
Fig. 2. A glial cell process (blue) enwrapping a synapse between a granule cell axon (the parallel fiber) terminal (uncolored) and a dendritic spines of Purkinje cell (light brown) in the cerebellar cortex. The black dots are SEPT5, which accumulates beneath the plasma membrane.
References
1. Ageta-Ishihara N. et al. Cell Rep. 6, 115352 (2025)
2. Scala S., Nishikawa M. et al. Brain 145, 3308 (2022)
3. Ageta-Ishihara N. et al. Nature Commun. 6, 10090 (2015)
4. Ageta-Ishihara N. et al. Nature Commun. 4, 2532 (2013)
5. Ihara M. et al. Neuron 53, 519 (2007)
Member
-

Professor
Makoto Kinoshita
kinoshita.makoto.u4(at)f.mail.nagoya-u.ac.jp
Physiological and pathological roles of the septin cytoskeletal system
-

Assistant Professor
Masashi Nishikawa
nishikawa.masashi.z7(at)f.mail.nagoya-u.ac.jp
Molecular pathophysiology of neurodevelopmental and neuropsychiatric disorders
-

Assistant Professor
Chikako Nakajima
nakajima.chikako.g7(at)f.mail.nagoya-u.ac.jp
Molecular mechanisms leading to brain functional regeneration
Group of Molecular and Cell Biology
Elucidation of the protein stability control system through ubiquitin
In our bodies, proteins are synthesized when they are needed and broken down when they are finished. Traditionally, proteins are strictly controlled during their synthesis process, and the degradation process is considered to be a mere garbage disposal mechanism, even though it is no longer needed in the cell. However, recent research has revealed that proteolysis is actually a regulatory system that actively controls various biological functions, and it is attracting a great deal of interest. Among them, we are focusing on and studying biological phenomena controlled by protein degradation via the ubiquitin-proteasome system.
Elucidation of biological phenomena controlled by the ubiquitin system
We have discovered that active proteolysis by the ubiquitin system plays an important role in many aspects of life phenomena. For example, we found that when the quantity and quality of nutrient sources available to cells change, proteins, which are switches for substance metabolism, are actively degraded to maintain the nutritional balance in cells. We have clarified that protein degradation by the ubiquitin system is important in order to respond flexibly to various environmental stresses that attack cells. In addition, when cells proliferate, organelles such as mitochondria are distributed to new cells (Fig. 1 left), and we found that the ubiquitin system plays an important role in this process as well. In this way, we are discovering new life phenomena controlled by the ubiquitin system and elucidating their mechanisms.
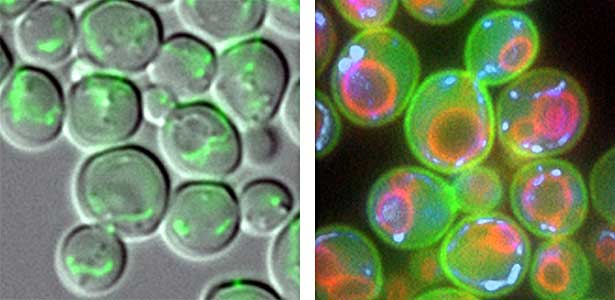
Fig. 1. Mitochondria passed to new cells (green)(left). Yeast cell membrane (green), vacuolar membrane (red), and mitochondrial DNA (blue) (right).
Study of the control of degradation of anthropogenic proteins through the ubiquitin system
The cell, which is the basic unit of life, is a closed space surrounded by a cell membrane (Fig.1 right). By closing the space of the cell membrane, the substances necessary for vital activity are concentrated without diffusion. It is no exaggeration to say that it is this work that brought about the birth of life. However, biological membranes not only act as “partitions” but also have many functions, such as the exchange of substances and information inside and outside the cell, the scaffold for chemical reactions that are indispensable for sustaining life, and the interaction with other cells. In many situations, the ubiquitin system is involved as an important mechanism. We are studying the origins and various functions of biological membranes such as cell membranes.
Study of the control of degradation of anthropogenic proteins through the ubiquitin system
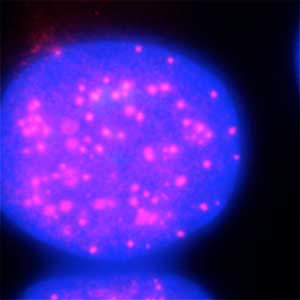
The ubiquitin system is responsible for the degradation of various proteins as a mechanism for protein degradation in cells. Using this system, we have developed an auxin-inducible degron (AID) method that breaks down proteins that we want to destroy using a substance called auxin, which is a plant hormone (Fig. 2). We are improving this proteolytic system so that it can be used more easily, and we are analyzing the function of target proteins using this proteolytic system.
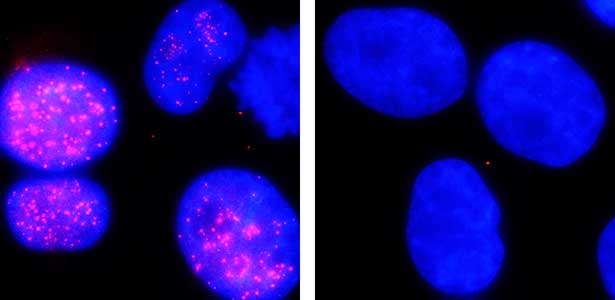
Fig. 2. Degradation of centromere protein (red) in the nucleus of human cells by auxin: before auxin treatment (left) and after auxin treatment (right).
References
1. Obara K. et al. Nature Commun. 13, 2005 (2022)
2. Nishimura K. et al. Nucleic Acids Res. gkaa748 (2020)
3. Nakatsukasa K. et al. Mol. Cell 59, 22 (2015)
4. Kamura T. et al. Science 284, 657 (1999)
Member
-

Professor
Takumi Kamura
kamura.takumi.k1(at)f.mail.nagoya-u.ac.jp
Elucidation of biological phenomena controlled by the ubiquitin system
-

Associate professor
Keisuke Ohara
obara.keisuke.r2(at)f.mail.nagoya-u.ac.jp
Study of the mechanism of maintenance of homeostasis of biological membranes by the ubiquitin system
-

Lecturer
Kohei Nishimura
nishimura.kohei.x8(at)f.mail.nagoya-u.ac.jp
Study of the control of degradation of anthropogenic proteins through the ubiquitin system

Group of Gene Expression and Regulation
Defining translated regions to understand genes
DNA is transcribed into mRNA, and mRNA is translated into proteins. However, identifying which regions of mRNA are translated is far more challenging than we expect. Utilizing cutting-edge technologies, we have discovered numerous novel translated regions, including those derived from non-coding RNAs, which were previously thought not to be translated into proteins. Furthermore, contrary to the conventional belief that a single mRNA produces only one protein in eukaryotic cells, multiple proteins can be translated from a single mRNA. Since gene names are typically assigned based on protein function, this discovery calls for a reassessment of current gene-naming conventions. By employing human iPS cells and genetically modified mice, we aim to deepen our understanding of processes related to the nervous system, immunity, cancer, and aging. This research has the potential to contribute to the development of novel disease treatments.
Discovery of "twin" genes required for sperm development
We discovered that Gm9999, a noncoding RNA expressed in sperm, encodes two small proteins, which we named Kastor and Polluks derived from the names of stars in the constellation Gemini (Fig. 1, left). The male mice lacking Kastor and Polluks were infertile. Kastor and Polluks were localized in the mitochondria of sperm, and electron microscopy revealed that sperm from mice lacking Kastor and Polluks had an abnormal mitochondrial morphology (Fig. 1, right). The small twin genes are essential for normal sperm development and may be one of the causes of male infertility in Human.
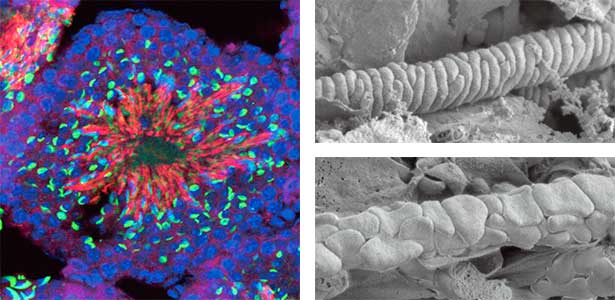
Fig. 1. Expression of Polluks proteins in mouse testis (left). Nuclei of all cells (blue), sperm heads (green), and Polluks proteins localized in sperm mitochondria (red). Scanning electron microscope images of sperm (right). Mitochondria of normal sperm (upper right) and those of sperm lacking the twin gene (lower right).
Discovery of Myo-ribosome essential for heart function
The ribosome, which is responsible for protein translation, is composed of about 80 protein subunits. We discovered that the heart has a specialized ribosome in which RPL3, one of the ribosome subunits, is replaced by RPL3L. Echocardiography of mice lacking the Myo-ribosome revealed reduced contractility of the heart (Fig. 2). Given that genetic mutations of RPL3L have been reported in patients with dilated cardiomyopathy, research on the Myo-ribosome may lead to the development of a new therapeutic approach for these patients.
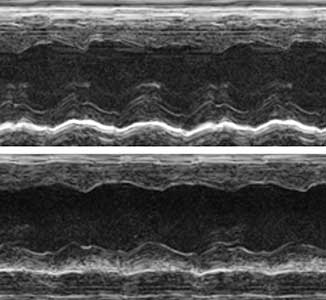
Fig. 2. Echocardiography of mice. Normal heart (upper) and heart deficient in Myo-ribosome (lower).
References
1. Shiraishi C. et al. Nature Commun. 14, 2131 (2023)
2. Kito Y. et al. EMBO J. e112869 (2023)
3. Mise S. et al. Nature Commun. 13, 1071 (2022)
4. Nita A. et al. PLoS Genet. 17, e1009686 (2021)
5. Ichihara K. et al. Nucleic Acids Res. 49, 7298 (2021)
6. Matsumoto A. et al. Nature 541, 228 (2017)
Member
-

Professor
Akinobu Matsumoto
matsumoto.akinobu.i3(at)f.mail.nagoya-u.ac.jp
Elucidating the unknown mechanism of central dogma
-

Assistant Professor
Kazuya Ichihara
ichihara.kazuya.d5(at)f.mail.nagoya-u.ac.jp
Understanding the regulatory mechanisms of diverse gene expression
Understanding of cytoskeleton dynamics from protein structures
Actin, a protein approximately 5 nm in diameter, polymerizes into filaments composed of two strands, playing numerous essential roles for the survival of eukaryotes. In fact, a significant portion (likely more than half) of the processes that generate force in eukaryotic cells rely on actin. The importance of actin is highlighted by the fact that the most abundant isoforms of actin in animals, skeletal muscle α-actin and cytoplasmic β-actin, remain unchanged between chickens and humans. Actin filaments exhibit high dynamism in the cells, undergoing continuous assembly and disassembly, a process crucial for their cellular functions. We conduct structural studies on the cytoskeleton, focusing on elucidating the molecular structure related to the actin filament dynamics through collaboration with numerous research groups.
Structural understanding of actin filament dynamics
High-resolution structural analysis is vital for understanding the assembly and disassembly of actin filaments at a structural level. In cells, older actin filaments are preferentially disassembled. Using cryo-electron microscopy and X-ray crystallography, we have clarified how actin filaments become aged and more prone to disassembly. Furthermore, the lifespan of the actin filaments is extended when they are pulled or twisted, and the mechanism behind this has been understood through mutant experiments and analysis of filament fluctuations using electron microscopy on actin filaments that are actually pulled or twisted.
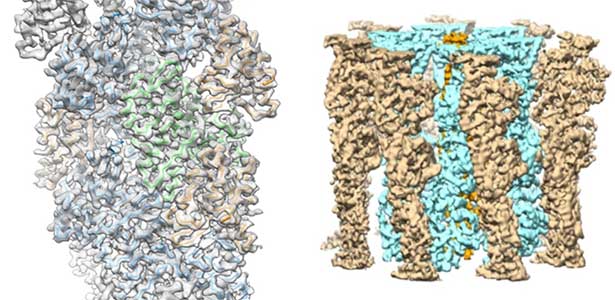
Fig. 1. Actin-cofilin complex structure (left) and ParM filament structure from Clostridium botulinum (right).
Cytoskeleton of bacteria and archaea
Actin filaments and microtubules, the main components of the cytoskeleton, are unique to eukaryotes, yet similar families exist in bacteria and archaea. For many years, we have studied ParM, a bacterial actin family protein, in collaboration with the Robert Robinson group. Although ParM subunits are structurally similar, they assemble into structurally diverse filaments, serving as a model for studying protein filament dynamics. In 2019, we elucidated that the ParM of Clostridium botulinum forms a complex structure consisting of 15 strands (Fig. 1, right). It is surprising that such a structure can be formed by the assembly of just one protein. Additionally, recent collaborative work with the Robinson group has enabled us to successfully analyze the structure of archaeal microtubules. While individual subunits of archaeal microtubules closely resemble those of eukaryotes, their filament structure significantly differs. Our goal is to elucidate the evolutionary transformation of these archaeal microtubules into eukaryotic microtubules through the course of evolution from archaea to eukaryotes.
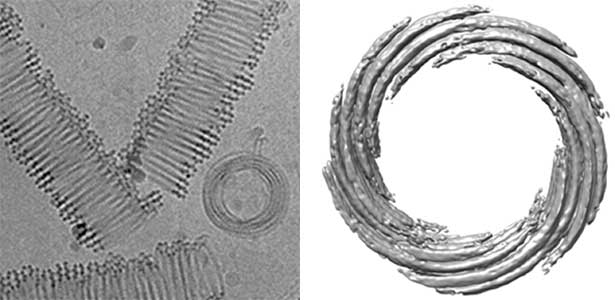
Fig. 2. Cryo-electron micrograph of archaeal microtubules (left) and its three-dimensional structure (right).
References
1. Okura K. et al. J. Mol. Biol. 435, 168295 (2023)
2. Kanematsu Y. et al. PNAS 119, e2122641119 (2022)
3. Akil C. et al. Sci Adv. 8, eabm2225 (2022)
4. Matsuzaki M. et al. Biomolecules 10, 736 (2020)
5. Mizuno H. et al. PNAS 115, E5000 (2018)
6. Tanaka K. et al. Nature Commun. 9, 1860 (2018)
7. Koh F. et al. Nature Commun. 10, 2856 (2019)
Member
-

Associate Professor
Akihiro Narita
narita.akihiro.x8(at)f.mail.nagoya-u.ac.jp

Group of Interdisciplinary Biology
Toward a quantitative understanding of biological phenomenon
Our ultimate research goal is "to quantitatively understand phenomena from the development of life to death." The core elements of our research include mathematical models, computer simulations, and AI technology. we consistently engage in innovative research by establishing collaborative networks with various universities, research institutes, and laboratories worldwide, integrating insights from diverse fields. Our specific focus lies in investigating the effects of pathogen infection and immune dysregulation. In particular we aim to unveil the maintenance and changes of tissues in vivo at the individual level through comprehensive analyses of time-course changes from gene expression to cellular function. As the first laboratory for interdisciplinary biology research in Japan, interdisciplinary Biology Laboratory, iBLab, stands at the forefront of advancing next-generation life science research driven by the innovative application of data science technology.
What is “interdisciplinary biology” in iBLab?
In our laboratory, we conduct groundbreaking life science research, combining cutting-edge model-driven approaches with emerging data-driven approaches to bridge diverse research fields. The core of our research methodology is creating a comprehensive approach that maximizes the strengths of each method and effectively compensates for their respective weaknesses. For example, by combining different types of data, we can carry out research that can quantitatively and accurately predict disease progression or treatment outcomes at an individual level. Moreover, our investigations extend to the application of insights derived from the digital realm to real-world scenarios. This is accomplished by generating extensive virtual data that replicates the characteristics of real-world data, allowing for the evaluation of diverse scenarios. Our research delves into these themes by seamlessly transitioning between the physical and digital realms (see Fig. 1).
Fig. 1. Research that integrates various fields.
The next generation of infectious disease research
In recent years, various infectious diseases such as COVID-19 and Mpox have spread globally and been successively introduced into Japan. The risk of importing these emerging and re-emerging infectious diseases is increasing due to global outbreaks of viral hemorrhagic fevers and zoonotic diseases like Ebola, Lassa fever, and Crimean-Congo fever. The COVID-19 pandemic has left a significant impact on the world, yet it has also generated an unprecedented volume of high-quality data, unparalleled in human history. To prepare for a potential future pandemic, it is crucial to analyze these datasets. Our group is committed to establishing an interdisciplinary platform using the latest data science technology to robustly support drug and vaccine development, as well as the formulation of appropriate countermeasures against infectious diseases and treatment strategies for severe cases.

Fig. 2. Infectious disease study using mathematical models, computer simulations, and AI technologies.
References
1. Hart W.S., Park H., Jeong Y.D. et al. PNAS 120, e2305451120 (2023)
2. Jeong Y.D., Ejima K., Kim K.S. et al. Nature Commun. 13, 4910 (2022)
3. Jeong Y.D., Ejima K., Kim K.S. et al. eLife 10, e69340 (2021)
4. Kim K.S., Ejima K., Iwanami S. et al. PLoS Biol. 19, e3001128 (2021)
5. Iwanami S., Ejima K. et al. PLoS Med. 18, e1003660 (2021)
Member
-

Professor
Shingo Iwami
iwami.iblab(at)bio.nagoya-u.ac.jp
Interdisciplinary research in the life sciences using mathematical science
-

Lecturer
Shoya Iwanami
iwanami.iblab(at)bio.nagoya-u.ac.jp
Quantitative understanding of life dynamics
Group of Genetics
Mechanism of cell-cell communications regulating tissue growth and homeostasis
Cells that constitute multicellular organisms, similar to individual organisms on Earth, coordinate with each other and also compete for survival. It has become apparent that such cell-to-cell communications play a crucial role in the morphogenesis and homeostasis of tissues and organs, as well as in the cancer development. In our laboratory, we utilize the Drosophila melanogaster, an excellent model system for analyzing these phenomena at the organismal level, focusing on intercellular communications. Through genetic approaches, live imaging, and the development of mathematical and physical models, we aim to elucidate the mechanism by which intercellular communication mediates (i) tissue growth and morphogenesis, (ii) the maintenance of tissue homeostasis, and (iii) the initiation and progression of cancer.
The mechanisms and physiological role of “Cell competition”
The phenomenon of “Cell competition”, where cells compete for survival, is increasingly recognized as playing a significant role in tissue growth, homeostasis, and cancer development. Our goal is to elucidate the mechanism by which cells recognize each other using chemical and mechanical signals, thereby determining ‘losers’ and ‘winners’. Furthermore, we are exploring the mechanism by which ‘winners’ expel ‘losers’ from the tissue. Simultaneously, we are aiming to reveal new physiological functions associated with this process.
Epithelial deformation through cell-cell communications
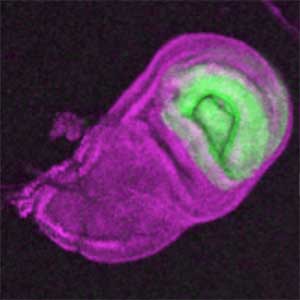
The diverse external morphologies of insects such as wings, legs, and exoskeletal horns are shaped through the three-dimensional deformation of the adult primordium, which is a ‘folded epithelial sheet’ structure. By integrating experimental approaches with mathematical and physical modeling, we aim to elucidate the fundamental principles governing the deformation of these folded epithelia (Fig. 1).
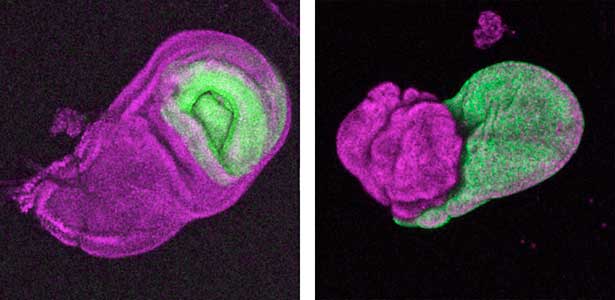
Fig. 1. “Folded” wing imaginal disc (left) and the adult wing (right).
Tumor development and progression through cell-cell communications
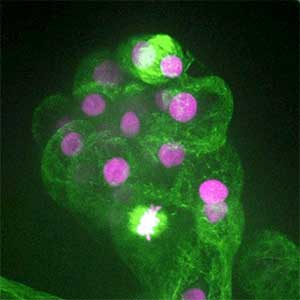
It has become apparent that the interaction between oncogenic cells and the surrounding cells plays a crucial role in the development and progression of cancer. We utilize a Drosophila tumor malignancy model to analyze the mechanisms behind tumor development and progression, focusing on the interactions between oncogenic cells and neighboring normal cells, such as epithelial cells and immune cells, as well as adjacent normal tissues (Fig. 2).
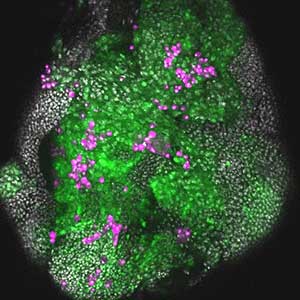
Fig. 2. The eye-antennal imaginal disc bearing oncogenic mutant cells (green). Immune cells (magenta) exhibit a substantial accumulation on the oncogenic mutant cells.
References
1. Akai N. et al. PLoS. Genet. 17, e1009300 (2021)
2. Ohsawa S. et al., Dev. Cell 44, 284 (2018)
3. Yamamoto M. et al. Nature 542, 246 (2017)
4. Ohsawa S. et al. Nature 490, 547 (2012)
5. Ohsawa S. et al. Dev. Cell 20, 315 (2011)
Member
-

Professor
Shizue Ohsawa
ohsawa.shizue.x5(at)f.mail.nagoya-u.ac.jp
Genetic dissection of the mechanisms underlying tissue growth/morphogenesis
-

Assistant Professor
Keisuke Ikawa
ikawa.keisuke.w6(at)f.mail.nagoya-u.ac.jp
Genetic dissection of mechanical cell competition
-

Assistant Professor
Yoshimasa Yagi
yagi.yoshimasa.j2(at)f.mail.nagoya-u.ac.jp
Non-cell-autonomous growth control by immune cells and analysis of its physiological functions.
-

Designated Assistant Professor (Young Researcher Unit)
Haruka Hiraoka
hiraoka.haruka.i9(at)f.mail.nagoya-u.ac.jp
Elucidation of the mechanisms of cancer progression driven by cell competition.
Group of Signaling Mechanisms
Elucidation of signal transduction mechanisms in animals
Organisms receive and respond to various stimuli (signals) from their internal and external environments. The mechanisms that link such signals to biological phenomena are called signal transduction mechanisms and are considered essential for understanding biological responses. In our laboratory, we study the signal transduction mechanisms that control the regeneration of severed nerves and neurodegenerative diseases using worm, fish, and mammalian cells to elucidate the control mechanisms of biological phenomena in vivo.
Signaling mechanisms regulating nerve axon regeneration
The nematode C. elegans is a 1-2 mm long worm that feeds on soil bacteria. It is an excellent model animal that has been in the spotlight in recent years because it has many genes homologous to those of mammals, including humans, and is easy to study genetically and molecularly. In our lab, we mainly study the signaling mechanisms that control axon regeneration in C. elegans. In particular, we aim to understand the role of signaling mechanisms conserved from worms to humans in regulating the regeneration of severed nerves in order to provide a new basis for the study of therapies for nerve injury (Fig. 1).
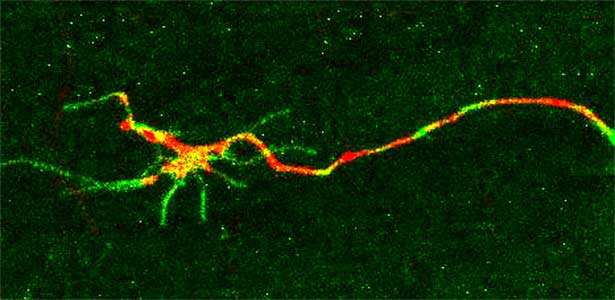
Fig. 1. Regenerating nerve axon. The severed nerve is visualized with red fluorescent protein and the collagen receptor DDR-2 protein is visualized with green fluorescent protein.
α-Synuclein aggregation mechanisms and synaptic degeneration
Parkinson’s disease is a neurodegenerative disorder characterized by the accumulation and propagation of α-Synuclein in the brain. This process begins as early as 10 to 20 years before symptoms appear (prodromal stage), and leads to progressive neural synapse dysfunction. Research is currently investigating the mechanisms of α-Synuclein aggregation and propagation during the prodromal phase of the diseases, as well as the synaptic degeneration caused by these processes at the molecular and cellular levels. The study is utilizing primary cultured hippocampal neurons and glial cells (Fig. 2).
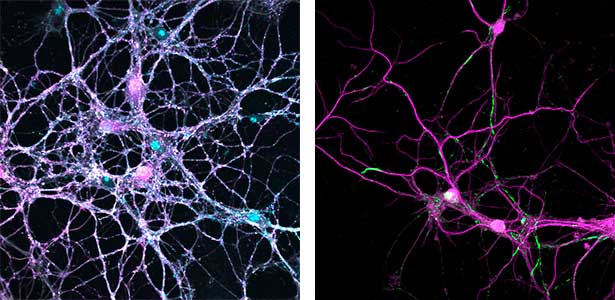
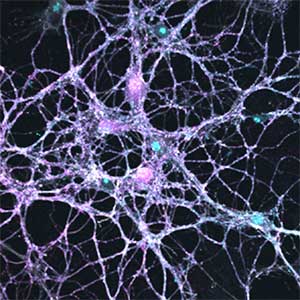
Fig. 2. Neuron-astrocyte co-culture (left) and α-Synuclein aggregates (right). Lewy neurite-like aggregates (rod-like structures) and Lewy body-like aggregates (spherical structures) of α-Synuclein (green) are formed in primary cultured neurons derived from mouse hippocampus (right).
References
1. Sakai Y. et al. EMBO Rep. 23, e55076 (2022)
2. Nakano S. et al. PNAS 117, 1638 (2020)
3. Hisamoto N. et al. Nature Commun. 9, 3099 (2018)
4. Alam T. et al. Nature Commun. 7, 10388 (2016)
5. Hanafusa H. et al. Nature Cell Biol. 17, 1024 (2015)
6. Li C., Hisamoto N. et al. Nature Neurosci. 15, 551 (2012)
Member
-

Professor
Naoki Hisamoto
i45556a(at)cc.nagoya-u.ac.jp
Analysis of signal transduction mechanisms using C. elegans as a model
-

Associate Professor
Hiroshi Hanafusa
q47371a(at)cc.nagoya-u.ac.jp
Analysis of signal transduction mechanisms in mammalian cells
-

Lecturer
Shunji Nakano
z48329a(at)nucc.cc.nagoya-u.ac.jp
Analysis of neural function using C. elegans as a model
-

Assistant Professor
Tsubasa Itou
itou.tsubasa.j0(at)f.mail.nagoya-u.ac.jp
Analysis of biological control mechanisms using fish as a model
Group of Development and Growth Regulation
Elucidation of the mechanism of cell morphology formation
We are conducting research aimed at understanding the chloroplast function involved in the development and growth of plants. Additionally, we are interested in the cellular mechanisms determining sperm length and their evolutionary background. Specifically, we are researching the elucidation of chloroplast function involved in plant morphogenesis and cell proliferation using molecular biology and cell biology techniques, focusing on the model plant Arabidopsis thaliana. Furthermore, we are developing genome editing techniques for the orchid species Phalaenopsis aphrodite. The fruit fly, Drosophila melanogaster, commonly used in genetic experiments, is a small and inconspicuous insect with few morphological features. However, interestingly, its sperm are exceptionally long, about 40 times longer than those of humans or mice, and there are even species closely related to it where the length of the sperm exceeds 5 cm. The extreme evolution of traits dependent on sex, such as the length of sperm, is explained by Ronald Fisher's "runaway evolution theory." We are interested in the cellular mechanisms determining such sperm length and their evolutionary background.
Chloroplast function involved in plant morphogenesis and cell proliferation
Chloroplasts not only conduct photosynthesis but also synthesize essential substances for plant survival, such as plant hormones, amino acids, and lipids. Additionally, they are involved in the regulation of nuclear genome gene expression, plant morphogenesis, and cell proliferation. We have discovered chloroplast-localized proteins that, when their function is impaired, lead to the emergence of cells without chloroplasts and abnormal leaf shapes or plant growth (Fig. 1). By conducting functional analyses of these proteins, we aim to elucidate chloroplast functions involved in plant morphogenesis and cell proliferation. Furthermore, we are working on the development of genome editing techniques for the orchid species and are also engaged in breeding new varieties of P. aphrodite.
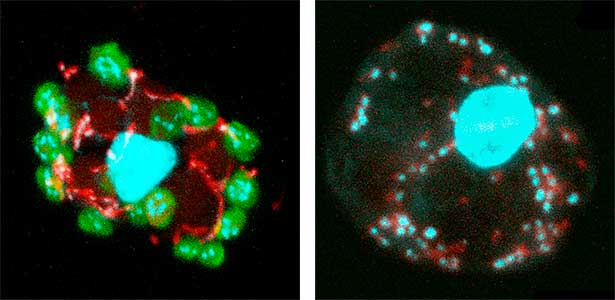
Fig. 1. Nucleus, chloroplasts (green), mitochondria (red), and DNA (blue) in leaf mesophyll cells of wild-type Arabidopsis thaliana (left) and crl mutant (right).
The role of giant mitochondria
Another characteristic of Drosophila sperm is the enlargement of mitochondria. Shortly after meiosis, the mitochondria, which were dispersed throughout the cell until recently, aggregate and become spherical (Fig. 2). These mitochondria have a diameter of approximately 6 micrometers, larger than those found in typical cells, and can be described as “giant” mitochondria. Over time, the shape of these mitochondria changes to elongated forms, pushing and stretching the cell from the inside, reaching lengths of up to 1.8 mm in D. melanogaster. Interestingly, a strong correlation has been found between the volume of giant mitochondria and the extreme length of the sperm. This suggests that the size of giant mitochondria may determine sperm length. We are aiming to prove this causal relationship and elucidate the evolutionary background of this intriguing phenomenon.
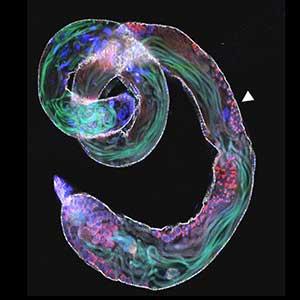
Fig. 2. Giant mitochondria (indicated by arrowheads) greatly enlarged and developed, identifiable even in photographs of the entire testis. The green structures are bundles of elongated sperm.
References
1. Semiarti E. et al. Indonesian J. Biotech. 25, 61 (2020)
2. Hudik E. et al. Plant Physiol. 166, 152 (2014)
3. Wang S.W. T. et al. Plant Cell Environ. 37, 2201 (2014)
4. Liu Z. et al. Dev. Growth Differ. 53, 822 (2011)
5. Sugiyama S. et al. Genetics 178, 927 (2007)
Member
-

Associate professor
Yasushi Yoshioka
yoshioka.yasushi.k4(at)f.mail.nagoya-u.ac.jp
Elucidation of organelle functions involved in plant morphogenesis and cell proliferation
-

Lecturer
Shin Sugiyama
ssugiya(at)bio.nagoya-u.ac.jp
The relationship between elongated sperm and giant mitochondria
Group of Intracellular Dynamics
Molecular mechanisms of intracellular motility in plant cells
In plants, where cells are surrounded by a rigid cell wall, the positional relationship between daughter cells generated by cell division is permanently maintained. Plants utilize phytohormones and secrete signal peptides to establish the somatic axis and regulate stem cells, which enables the formation and organization of organs such as flowers and leaves. Our research group studies the molecular mechanisms underlying these morphogenetic processes using the moss Physcomitrium patens. P. patens is amenable to techniques such as high-resolution imaging, CRISPR/Cas9 genome editing, and inducible RNAi, making it a particularly suitable model plant for cell biological analyses. By taking advantage of the moss’s unique properties, we are currently focusing on the cytokinesis process of cell division in plants.
Cell division in plants
One of our main interests lies in how plant cells divide, a process called cytokinesis. During this process, plants partition a mother cell by forming the cell plate between two daughter nuclei. This unique cytokinetic system is widely conserved among land plants. However, because of its uniqueness, there are still many mysteries about plant cell division, such as how the cell plate is positioned and how it is formed. We aim to uncover the molecular mechanisms underlying plant cell division by focusing on proteins involved in cell plate formation.
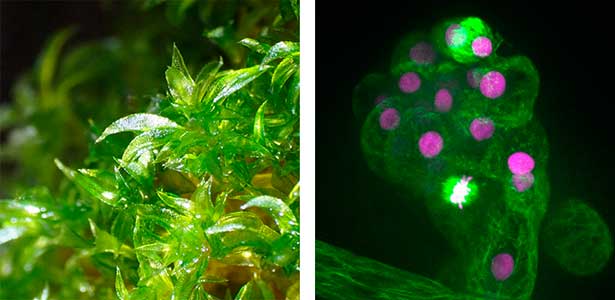
Fig. 1. Gametophores in P. patens and the early gametophore visualized by microtubules (green) and DNA (magenta).
References
1. Yamada M. et al. Nature Plants 11, 340 (2025)
2. Leong S.Y. et al. Plant Cell 32, 683 (2020)
3. Wu S.Z. et al. Biophys. Rev. 10, 1683 (2018)
4. Yamada M. and Goshima G. Plant Cell 30, 1496 (2018)
5. Yamada M. et al. J. Cell Biol. 216, 1705 (2017)
Member
-

Assistant Professor
Moe Yamada
Yamada.moe.d0(at)f.mail.nagoya-u.ac.jp
Mechanism of cell division in moss
Cell division, known as mitosis, serves as a fundamental process for growth and development across all multicellular organisms. Plant mitosis exhibits several distinctive features that set it apart from animal cell division. Our main research theme is to discover plant-specific molecules and pathways involved in cell division. Our main model organism is the bryophyte Physcomitrium patens (Physcomitrella). As an experimental organism, it has the nickname "green yeast" because of its high DNA homologous recombination rate and almost haploid life cycle, making it an excellent choice for genetic screening and cell biology research. In addition to basic research, we have a strong interest in the development of new technologies in plant cell biology, such as microfluidic devices.
Unique features of plant cell division
Plant cell division, while sharing the main stages of eukaryotic mitosis, is distinguished from animal mitosis by lack of centrosomes, preprophase band, cytokinesis via phragmoplast. We work to discover unique plant-specific molecules and pathways involved into cell division and potentially harness those towards practical applications. There are several ongoing projects, including CRISPR/Cas9 genetic screening to discover novel cell division genes, investigating relationship between plant kinetochore and polyploidy and mechanisms of spindle positioning in plants (Fig. 1 left).

Fig. 1. Colony of moss Physcomitrium patens (left). Dividing moss cell expressing microtubule (magenta) and kinetochore (green) markers (right).
Microfluidic Device for high resolution cytoskeleton imaging
Microfluidic chip or device is a set of micro-channels molded into a polymer material (PDMS) in which plant cells can be grown without any adverse effects. We can precisely modulate the channel environment by controlling liquid flow in the channels, introducing or wash-out chemical compounds, and observing the cellular response. Furthermore, microdevice offers excellent properties for high-resolution imaging and can be used to visualize protein dynamics at the molecular level. We use microdevices for high-resolution cytoskeleton imaging in the cells of moss P. patens and testing the effect of various chemical compounds on cytoskeleton dynamics (Fig. 1 right).
References
1. Yoshida M.W. and Kozgunova E. Methods Mol. Biol. 2604, 143-158 (2023)
2. Kozgunova E. et al. Nature Commun. 13, 2488 (2022)
3. Kozgunova E. et al. eLife 8, e43652 (2019)
4. Kozgunova E. et al. Sci. Rep. 9, 15182 (2019)
5. Kozgunova E. et al. Plant Cell Physiol. 57, 848 (2016)
Member
-

YLC Designated Assistant Professor
Elena Kozgunova
kozgunova.elena.b1(at)f.mail.nagoya-u.ac.jp
Research on dissociation mechanism of cell-cell adhesion
White blood cells and metastatic cancer cells are able to open the walls of blood vessels, but the mechanism by which these cells dissociate the tight bonds between blood vessel cells is still unknown. If we uncover this mechanism, we may be able to create drugs for inflammatory diseases and cancer metastasis. We are attempting to solve this mystery, paying attention to hemorrhage mechanism by snake toxins.
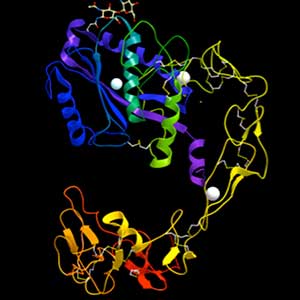
Fig. 1. Structure of ADAM-type haemorrhagic toxin.
References
1. Seo T. et al. FEBS J. 284, 1657 (2017)
Member
-

Associate Professor
Satohiko Araki
araki.satohiko.u0(at)f.mail.nagoya-u.ac.jp
Group of Cellular and Membrane Biology
Studies of cell adhesion function
Multicellular organisms are formed when a large number of cells of different types come together and attach to each other. It is therefore clear that cell adhesion is an essential function for multicellular organisms. We focus mainly on hemidesmosomes, the cell-to-matrix adhesion machinery of epithelial tissues, and study them using cultured cells.
Hemidesmosomes attach epithelial tissue to the basement membrane
Epithelial tissue is a sheet-like structure that covers the body surface and lumen of multicellular organisms. Epithelial tissue protects the body of multicellular organisms from various external stresses, while at the same time mediating or restricting the exchange of substances between the inside and outside of the body. It corresponds to the cell membrane of single-celled organisms. In order for epithelial tissue to remain attached to the body, it must be firmly attached to an extracellular matrix structure called the basement membrane, which lies directly beneath the epithelial sheet. Hemidesmosomes are adhesion protein complexes that bind epithelial tissue to this basement membrane (Fig. 1, left). It is known that in patients with a congenital inability to form hemidesmosomes or an acquired disease that prevents the proper functioning of hemidesmosomes, the epidermis of the skin becomes detached from the basement membrane. We are investigating the mechanisms of hemidesmosome formation and disassembly, including their relationship to disease, mainly using cultured cells derived from epidermis (Fig. 1, right).
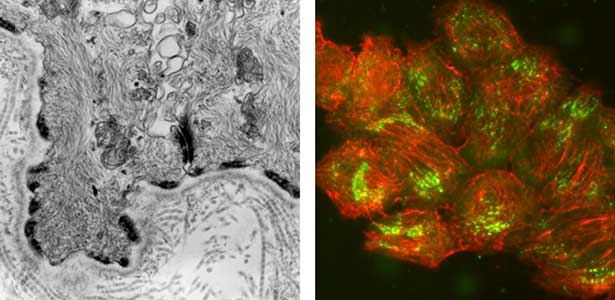
Fig. 1. Electron micrograph of esophageal epithelial basal cells (left). Electron-dense hemidesmosomal structures are localized at the basal side of the epithelial cells, facing the basement membrane. Visualisation of hemidesmosomes in cultured cells derived from rat epidermis (right). Hemidesmosomes (green) and keratin filaments (red) are shown.
References
1. Maglie R. et al. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 249
2. Hashimoto T. Hirako Y. and Tsuruta D. Exp. Deramtol. 25, 267, 2016
3. Yamauchi T. et al. J Dermatol. Sci. 76, 25 2014
4. Hirako Y. et al. Exp. Cell Res. 324, 172, 2014
Member
-

Lecturer
Yoshiaki Hirako
hirako.yoshiaki.i0(at)f.mail.nagoya-u.ac.jp
Elucidation of the functional regulation of the hemidesmosome, a cell-to-matrix adhesion protein complex
Studies of cell membrane dynamics
To understand the molecular mechanisms responsible for cell morphogenesis, maintaining the internal structures, and generating cellular movements, we utilize a reconstitution approach using the liposomes. Cell morphology and movement are under the highly orchestrated interactions of biological membranes with the cytoskeletal system, molecular motors, and various other regulation factors. By observing and analyzing their dynamics during interact with liposomes, we aim to understand the mechanism at the molecular level.
What is molecular mechanism responsible for the morphology, structure, and movement of cells?
Amphipathic phospholipids spontaneously assemble into bilayer membranes in aqueous solution and necessarily form liposomes, which are closed-membrane vesicles. Liposomes have been used as the most simplified models to study biological membranes. Real-time observation of liposomes using optical microscopes demonstrated that lipid membrane itself has the ability to cause large deformations that even involve topological changes such as fusion, fission, opening pore, or inside-out inversion (Fig. 1). In addition, it has been revealed that such dynamic behaviors of lipid membrane occur under the collaboration with various biological factors. Typical examples are macromolecules including proteins and nucleic acids, and cytoskeletons such as actin and/or molecular motors. Our group observes the phenomena that are induced when liposomes are exposed to such various factors that are candidates to be involved in the morphogenesis and movement of cells, and analyzes the dynamic mechanisms. In addition to the study for membrane dynamics, we have also progressed research focusing on the features of biological factors as natural soft matter. For examples, nematic liquid crystal formation of actin filaments and the dynamics of cytoskeletons, nucleic acids, or membranes when the liquid-liquid phase separation takes place (Fig. 2).
Fig. 1. Examples of dramatic deformations involving also topological changes of liposomes. From top to bottom: fusion, fission, perforation, front-to-back reversal. All dark-field microscopy images.
Fig. 2. Distribution in microdroplets observed when actin fibres (red) and DNA (green) are added simultaneously to a binary solution of polyethylene glycol and dextran in liquid-liquid phase separation.
References
1. Waizumi T. et al. J. Chem. Phys. 155, 075101 (2021)
2. Tanaka S. et al. Commun. Phys. 1, 18 (2018)
3. Tanaka-Takiguchi Y. et al. Langmuir 29, 328 (2013)
Member
-

Lecturer
Kingo Takiguchi
j46037a(at)nucc.cc.nagoya-u.ac.jp
Sugashima Marine Biological Laboratory

Group of Marine Biology
Cell biology of marine fungi and macroalgae
This research group has long studied intracellular dynamics, particularly processes involving microtubules, using model cell types derived from animals, plants, and fungi at the Higashiyama Campus in Nagoya (2007–2023). However, our focus has recently shifted to cell biology in marine organisms (2020–). In these non-model organisms, even fundamental cellular activities, such as cell growth and division, often exhibit different dynamics from those of model cells. We aim to elucidate the underlying mechanisms and the rationale. Unlike model organisms, for which various techniques have been established and experimental materials are easily available, we have to develop the experimental system from scratch, which is challenging but also a lot of fun. The research is conducted at Nagoya University’s Marine Biological Laboratory (NU-MBL) on Sugashima Island in Toba.
Marine fungi
Fungi, especially several model species like S. cerevisiae (budding yeast), S. pombe (fission yeast), and A. nidulans (filamentous fungus), have made significant contributions to the fields of biology and medicine, playing a crucial role in elucidating fundamental cellular mechanisms. However, recent research has unveiled a diverse range of fungal species inhabiting marine environments, many of which exhibit distinct cell growth and division patterns compared to model fungi. For example, several black yeast species collected at Sugashima MBL have shown the ability to adapt their growth and division strategies in response to environmental cues (a phenomenon known as phenotypic plasticity). We aim to conduct cutting-edge cell biology research on fungi originating from marine sources.
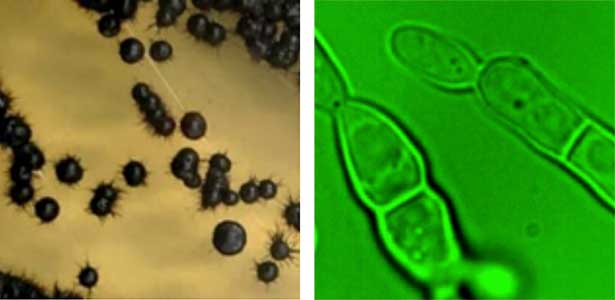
Fig. 1. Black yeast collected at Sugashima MBL (colonies and cells).
Marine macroalgae (seaweeds)
Sugashima is renowned for its diverse array of seaweeds, including nori (a seaweed commonly used in Japanese cuisine). Our research delves into the cellular intricacies of these seaweeds. Many macroalgal species exhibit cellular characteristics that markedly differ from those of terrestrial organisms. For example, while cell division is a fundamental process for all living organisms, seaweeds showcase unconventional patterns even in this activity, challenging conventional understanding. When seeking to understand these phenomena, it is not possible to directly apply insights gleaned from research on model land plants. Presently, our focus is centred on the study of the green alga Bryopsis. This macroalga, which grows to ~10 cm, resembling bird feathers, is remarkably a single-celled organism containing multiple nuclei (called coenocyte). We are intrigued by how it forms intricate shapes without undergoing cell division.

Fig. 2. Green feather alga Bryopsis sp. collected at Sugashima MBL (right; magnified image).
References
1. Yoshida M.W. et al. Nature Plants 9, 733 (2023)
2. Goshima G. Genes Cells 27, 124 (2022)
3. Shirae-Kurabayashi M. et al. PLoS ONE 17, e0264827 (2022)
4. Kim J. and Goshima G. PNAS 119, e2114429119 (2022)
5. Kozgunova E. et al. Nature Commun.13, 2488 (2022)
6. Tsuchiya K. and Goshima G. J. Cell. Biol. 220, e202104114 (2021)
Member
-

Professor
Gohta Goshima
Ggoshima(at)gmail.com
Diversity and evolution of marine organisms
The oceans, encompassing approximately 70% of Earth's surface, are inhabited by a plethora of organisms, ranging beyond the fish and algae consumed by humans, to a diverse array of life forms that interact with us both directly and indirectly. To sustain a symbiotic relationship with other life forms and continue to benefit from them, it is imperative to catalog the types of organisms present, their locations, and their populations. Our research primarily focuses on the systematics of marine invertebrates, exploring the biodiversity, morphological, and ecological evolution of these organisms. By collecting specimens and comparing their morphological and genetic characteristics to those already known, we identify and describe new species. Furthermore, we investigate the evolutionary pathways of organisms with unusual shapes or ecologies through detailed observations of their microstructures and ecological behaviors in controlled environments.
Systematics of marine invertebrates
The systematics of marine invertebrates is crucial due to their significant role in ecosystems, yet the field remains in a state of early development, with many species yet to be described. Our efforts span from the Arctic to the Antarctic, from intertidal zones to the hadal depths, utilizing research vessels and diving expeditions to collect and classify organisms from every corner of the globe. Despite the heavy reliance on fieldwork, substantial deskwork is also integral to our research process. When a collected specimen does not match any previously described species, it is classified as a new species. The thrill of discovering unknown creatures based on historical records is akin to the excitement felt by Indiana Jones, making this field especially appealing to those who cherish fieldwork.
Morphological and ecological evolution of "strange" creatures
Exploring the morphological and ecological evolution of “strange” creatures raises questions like why the males of certain anglerfish are smaller, or why clownfish cohabitate with sea anemones. By elucidating their evolutionary history, we delve into the origins of such unusual traits. While our primary focus is on symbiotic ecologies and morphological evolution, our interests extend to any “strange” creatures, including the advanced evolution of eyes in annelids.

Fig. 1. Introduction to ‘strange’ organisms. Scaly worm (left), which has acquired a high-performance eye. Cirrus symbioses with octocorals (right).
The mechanisms behind the creation and maintenance of biodiversity
The mechanisms behind the creation and maintenance of biodiversity are exemplified by coral reefs, which support a vast array of life, contributing to their biodiversity. Organisms that play a pivotal role in shaping these ecosystems are known as ecosystem engineers. Recent diving expeditions around Sugashima have revealed the role of polychaetes as ecosystem engineers. Our ongoing research aims to understand the role of these “polychaete forests” within marine ecosystems, including the variety of organisms that utilize them.

Fig. 2. Lugworm forest consisting of arboreal nests. Replaces algal beds in deep reef zones and nurtures many organisms.
References
1. Jimi N. et al. Royal Soc. Open Sci. 10, 230039 (2023)
2. Jimi N. et al. Deep-Sea Res. Part I 196, 104025 (2023)
3. Jimi N. et al. Zoosystematics and Evolution 99, 149 (2023)
4. Jimi N. et al. Parasitol. Inter. 90, 1 (2022)
5. Jimi N. et al. J Zool Systema Evol. Res. 59, 801 (2021)
Member
-

Lecturer
Naoto Jimi
beniimo7010(at)gmail.com
Nagoya University Center for Gene Research
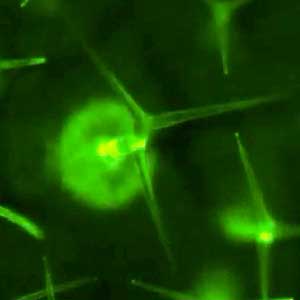
Group of Plant Molecular Signaling
Analysis of signal transduction mechanisms in plant immunity
As sessile organisms, plants do not induce acquired immunity like vertebrates. However, plants have established sophisticated defense networks through innate immunity, where they detect the invasion of pathogens through innate receptors, and their unique immune system. We aim to elucidate the signal transduction mechanisms that control plant immune networks.
Immune system controlled by plant hormones: Systemic acquired resistance
Plants possess innate immunity as a defense system. Upon perception of microbe-associated molecular patterns (MAMPs), such as microbial cell walls and flagellar components as well as toxic effectors secreted by pathogens, through specific receptors, diverse signal transduction pathways are activated to stimulate the immune system. Induction of local innate immunity, plants release long-distance signals from infected leaves, leading to the activation of the immune system even in non-infected leaves. This plant-specific immunity is known as systemic acquired resistance (SAR). In SAR, salicylic acid (SA) accumulates in non-infected leaves and induces SA-responsive genes. We aim to elucidate the signaling pathway of SAR, a key immune system in plants.

Fig. 1. Mechanical stimuli initiates the concentric propagation of intercellular calcium waves away from trichomes on leaf surfaces. Ca2+ imaging using 35Spro:GCaMP3 (Col-0). The leaf surface of a 4-week-old plant was gently brushed.
Systemic acquired resistance and crosstalk with environmental signals
The successful activation of SAR depends not only on the plant’s immune capacity but also on diverse environmental factors such as temperature, light, water, and herbivores. Analyses of crosstalk systems between signaling pathways activated by environmental factors and immunity are challenging, and most of the network remains unclear. We developed a cell-free protein synthesis system that comprehensively synthesizes candidate regulators of each signaling pathway, and investigate their regulatory mechanisms in vitro and their roles in vivo. Currently, we are studying the crosstalk between 1) jasmonic acid signaling involved in resistance against herbivores and necrotrophs, 2) light signaling, and 3) abscisic acid signaling involved in drought stress responses.
Novel immune system induced by mechanical stimulation
It is estimated that there are several hundred types of receptors in each plant that recognize microorganisms, but it is uncertain whether they identify all potential pathogens in nature. Recently, we discovered that in plants, trichomes, which are hair-like structures on leaf surfaces, function as sensors for mechanical stimuli and temporarily activate the immune system by detecting stimuli such as rain (Fig. 1). Since many pathogens spread and infect through raindrops or wind, plants seem to perceive these mechanical stimuli as warning signals, preparing for potential infections. Currently, we are studying the downstream signaling mechanisms involved in the perception and signal transduction of mechanical stimuli (Fig. 2).
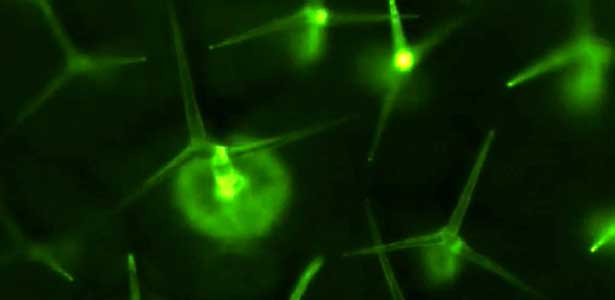
Fig. 2. Mechanosensory trichomes activate an immune response. Mechanosensory trichome cells initiate intercellular calcium waves in response to mechanostimulation. [Ca2+]cyt derepresses CAMTA and activates the phosphorylation of MPK3 and MPK6, thereby inducing WRKY-dependent transcription.
References
1. Matsumura M., Nomoto M. et al. Nature Commun. 13, 1216 (2022)
2. Nomoto M. et al. Cell Rep. 37, 110125 (2021)
3. Nomoto M. and Tada Y. Genes Cells 23, 46 (2018)
4. Spoel S.H. et al. Cell 137, 860 (2009)
5. Tada Y. et al. Science 321, 952 (2008)
Member
-

Professor
Yasuomi Tada
ytada(at)gene.nagoya-u.ac.jp
Regulatory mechanisms of environmental response by plant immunity
-

Lecturer
Mika Nomoto
nomoto(at)gene.nagoya-u.ac.jp
Regulation mechanisms of plant immunity by environmental signals
The way of life of unicellular microorganisms deciphered from genome information
There are a large number of microorganisms on the earth that are invisible to the naked eye, and they are adapted to various environments. They have survived by adapting to each environment that is very stressful for us in our normal lives: temperatures range from 0 to 130°C, pH from 0 to 13, and salinity from freshwater to saturated salinity. In my laboratory, we are approaching the diversity of adaptations to extreme environments resulting from evolution over a long period through mutagenesis and genome analysis. In particular, we use extremely halophilic archaea that prefer to grow in saturated salt environments as material to investigate the unique properties of these archaea.
Development of tools for microbial genome analysis
The advent of next-generation sequencing has made genome analysis relatively easy, and forward genetics, which works by creating mutants and searching for their causative genes, can be performed very efficiently. To effectively advance forward genetics, we have devised an efficient nucleic acid purification system using 96-well deep wells for concurrent culture and 96-well PCR plates and magnetic beads to efficiently advance mass genome analysis. In addition, we have established a data analysis workflow to process data from 300 to 1000 microbial genomic DNAs in a single analysis.
Extremely halophilic archaea
Extremely halophilic archaea (hereafter referred to as halobacteria) are a type of archaea that grow in a medium containing nearly saturated salt (25%) (Fig. 1, left). Most halophilic bacteria have a protein similar to rhodopsin, which is used in human vision. Some species of bacteria have only a chloride ion pump, called halorhodopsin (hR), and prefer to grow in alkaline environments with a pH of 9~11. hR is purple in the presence of chloride ions and blue in the absence of ions, and X-ray crystallography has revealed its conformational changes (Fig. 1, right). To perform this structural analysis, we used a mutant strain expressing a large amount of hR, and genomic analysis revealed a mutation (D324N) in the protein NP0654A. We expect that the mutation in NP0654A probably caused a significant increase in hR gene expression. Why not use the mutants to conduct a treasure hunt for genes that microorganisms possess?
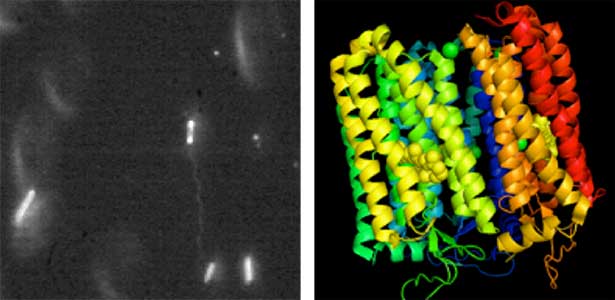
Fig. 1. Dark-field micrograph of an extremely halophilic archaeon (left) and the three-dimensional structure of halorhodopsin (right).
References
1. Ihara K. et al. FEBS Lett. 582, 2931, 2008
2. Kouyama T. et al. J.Mol.Biol. 396, 564, 2010
3. Tomatsu C. et al. FEBS Lett. 592, 1634, 2018
4. Kurauchi T, et al. FEMS Microbiol. Lett. 368, fnab070, 2021
Member
-

Associate Professor
Kunio Ihara
ihara.kunio.z9(at)f.mail.nagoya-u.ac.jp
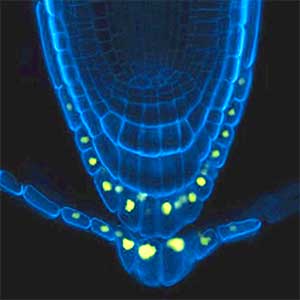
Group of Multicellular Coordination
Toward understanding multicellular organization in plants
Within the bodies of multicellular organisms, diverse groups of cells communicate and collaborate to carry out various activities. Consequently, the structure of the individual is elaborately established, enabling it to flexibly respond to environmental changes. We aim to unravel the mechanisms governing the establishment and maintenance of order among these cell groups and comprehend how this order dynamically adjusts to different situations. Specifically, we focus on plants, which demonstrate flexible adaptation in changing environments where they germinate. Our research encompasses the exploration of molecules conveying various information among cells, elucidation of the mechanisms involved in this information transmission, functional analysis of related genes, and attempts to uncover novel mechanisms for multicellular order by harnessing synthetic small compounds with unique properties.
Beautiful multicellular order inside plant bodies
Plants comprise a variety of cell types, spanning from small to large. These cells are meticulously arranged in aesthetically appealing patterns, like geometric expressions (Fig. 1, left). Such exquisitely ordered cell patterns can be abundantly observed throughout the plant body. We are captivated by the mechanisms responsible for generating these orderly and beautiful patterns within the plant body. Our focus lies in understanding the role of molecules, specifically signaling molecules, involved in transmitting information between cells. We emphasize the significant role played by signaling molecules that originate exclusively in specific cells within the plant body (Fig. 1, right).
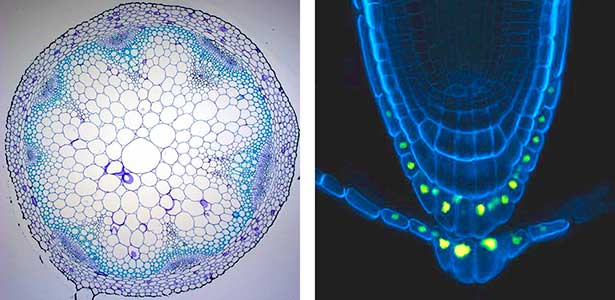
Fig. 1. Cross-sectional photograph of an Arabidopsis thaliana stem (Left). Cell walls are stained. Photograph at the root tip (Right). The nuclei of cells producing the signal molecules we focus on show yellow fluorescence protein. Cell walls are stained with blue fluorescence.
Intercellular communication governing an organism
Within the intercellular communication in the plant body, there exists information that can alter the fate of the entire body. For instance, a failure to transmit even a single piece of information can lead to rapid senescence and deterioration of the entire body (Fig. 2). We are interested in unraveling the mechanisms underpinning the well-regulated and organized order as an individual organism.

Fig. 2. A normal Arabidopsis thaliana (left) and an instance where the individual undergoes rapid senescence (right).
References
1. Murao M. et al. Plant Cell Physiol. 64, 1167 (2023)
2. Negoro S. et al. Plant Cell Environ. 46, 451 (2023)
3. Uchida N. et al. Nature Chem. Biol. 14, 299 (2018)
4. Hirakawa Y. et al. Nature Commun. 8, 14318 (2017)
5. Tameshige T. et al. Curr. Biol. 26, 2478 (2016)
Member
-

Professor
Naoyuki Uchida
uchinao(at)gene.nagoya-u.ac.jp
Formation, maintenance, and change of multicellular homeostasis in plant body
-

Assistant Professor
Asuka Higo
higo.asuka(at)gene.nagoya-u.ac.jp
Signals that orchestrate the growth of plant body
Institute of Transformative Bio-Molecules (ITbM)

Group of Plant Chemical Genetics
Chemical biology of parasitic plants
Parasitic plants are heterotrophic plants that grow by taking nutrients from other plants. In Africa, one of these parasitic plants called Striga hermonthica has been causing serious damage to crop production. In our laboratory, we take small-molecule approach to elucidate how parasitic plants evolved to communicate with host plants through signaling molecules. With the information obtained, we develop new methods to control Striga for securing food production in Africa.
The mechanism of germination by sensing host-exuded strigolactones
Unlike normal plants, seeds of Striga stay dormant and never germinate unless host plants growing around (Fig. 1). When host plants grow nearby, the seeds sense host-exuded strigolactones and initiate germination. We developed a fluorogenic probe called yoshimulactone green which reacts with strigolactone receptors and emits fluorescence. The probe enabled us to identify strigolactone receptors and visualized wave-like propagation of strigolactone perception dynamics within the seeds (Fig. 2). We are studying how this pattern is created and how this dynamics is linked to germination.

Fig. 1. Striga is a parasitic plant of maize with pink flowers.
Combatting against striga with suicide germination-inducing molecule
By identifying of strigolactone receptors along with the development of convenient tool to monitor their functions, it became possible to rationally develop synthetic agonists of strigolactone receptors. We succeeded in developing Sphinolactone-7, a hybrid molecule of synthetic molecule and natural strigolactones, which is extremely potent germination stimulant for Striga. This molecule led Striga seeds to germinate in the absence of host plants and let them die. We are currently testing the suicide germination method using sphinolactone-7 in Kenya.
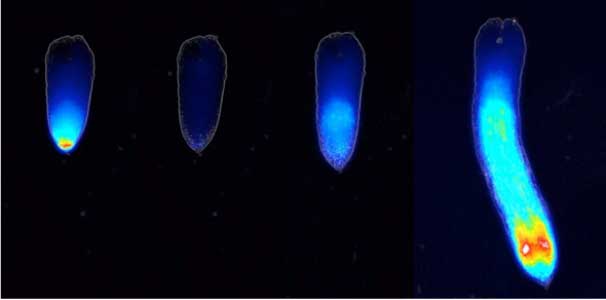
Fig. 2. Strigolactone recognition patterns occurring in waves in striga seeds (from left to right: before treatment, after 4 hours, after 11 hours, and after 27 hours).
References
1. Uraguchi D. et al. Science 362, 1301 (2018)
2. Tsuchiya Y. et al. Science 350, 203 (2015)
Member
-

Designated Professor
Yuichiro Tsuchiya
yuichiro(at)itbm.nagoya-u.ac.jp
Group of Molecular Chronobiology
Molecular dissection and regulation of the circadian clock
The circadian clock controls daily rhythms of various physiological processes including sleep/wake behavior and metabolism. Disruption of the circadian clock by shift work or genetic mutations can lead to diseases such as sleep disorders, metabolic diseases, and cancer. Therefore understanding the circadian clock system has major implications for improving human health. The circadian clock is composed of a transcriptional regulatory network of clock genes, but important questions remain, such as how it produces accurate rhythms and how it regulates physiological functions. Our group studies the molecular mechanisms of the circadian clock by combining biochemistry and chemical biology. The discovery of compounds that regulate the circadian clock could also lead to the development of therapeutic agents for diseases.
Discovery of compounds that regulate the circadian clock
To elucidate the molecular mechanisms of the circadian clock, we have been applying chemical biology approaches, which use chemical compounds to study the mechanisms of biological processes. Among tens of thousands of compounds with diverse structures, we identified several compounds that alter the circadian rhythm of human cells. From functional analysis of these new compounds, we discovered the first compounds that selectively act on each of the clock proteins CRY1 and CRY2, which constitute the circadian clock (Fig. 1). Furthermore, by analyzing the crystal structures of these compounds in complex with CRY proteins, we revealed the unique mechanism underlying the isoform selectivity. We are using our original compounds to uncover the mysteries of the circadian clock.

Fig. 1. Compounds selectively target each of the clock proteins CRY1 and CRY2.
Basic research for the treatment of diseases
In modern society, disturbances of the circadian clock and resulting various diseases are becoming increasingly serious problems. Compounds that regulate the circadian clock are expected to provide the starting point for drugs to treat these diseases. We have found that compounds targeting CRY inhibit hepatocyte glycogenesis, promote brown adipocyte differentiation, and inhibit glioblastoma stem cell proliferation. Furthermore, we found compounds that specifically inhibit CK2, a kinase associated with the circadian clock, and inhibit the proliferation of acute myeloid leukemia cells (Fig. 2). By analyzing the effects of these compounds on animal models of metabolic syndrome and cancer, we are also conducting basic research for the treatment of diseases.
Fig. 2. Compound specifically inhibits clock-related kinase CK2.
References
1. Miller et al. Proc. Natl. Acad. Sci. USA 119, e2203936119, 2022.
2. Miller, Srivastava et al. Proc. Natl. Acad. Sci. USA 118, e2026191118, 2021.
3. Miller, Son, Aikawa et al. Nature Chem. Biol. 16, 676-685, 2020.
4. Oshima, Niwa et al. Science Advances 5, eaau9060, 2019.
5. Hirota et al. Science 337, 1094-1097, 2012.
Member
-

Designated Associate Professor
Tsuyoshi Hirota
Thirota(at)itbm.nagoya-u.ac.jp
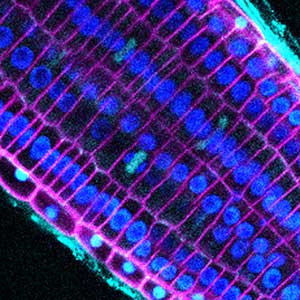
Group of Chemical Physiology and Advanced Imaging
Investigating plant physiological processes through live cell exploration
Land plants, as sessile organisms, have evolved remarkable adaptive capabilities to cope with their environments. Their ability to flexibly alter morphology in response to environmental conditions, and to optimize nutrient uptake from the soil, are recognized as key factors supporting their environmental adaptability. In our group, we address several fundamental questions in plant biology, such as the mechanisms underlying the establishment of cell polarity, the regulation of nutrient transporter dynamics, and the dynamic mechanisms by which plants respond to environmental stimuli. These investigations are primarily driven by live-cell imaging technologies. As members of the Institute of Transformative Bio-Molecules (ITbM), we conduct our research in close collaboration not only with researchers in the Department of Biological Science but also with chemists at ITbM and the Frommer group at Heinrich Heine University Düsseldorf (HHU) in Germany. Together, we are also engaged in the development of novel bioimaging tools that support our research endeavors.
Unraveling the functional aesthetics of plant cells
In plant cells, the optimal function of various membrane proteins—such as transporters for nutrients and phytohormones, and receptor-like kinases—is achieved through their asymmetric distribution along the cell surface (Fig. 1). This phenomenon, known as cell polarity, remains poorly understood in plants. Our research focuses on elucidating the mechanisms that regulate plant cell polarity by using membrane proteins such as the boron transporter BOR1, which localizes toward the inner tissues, and the boric acid channel NIP5;1, which localizes toward the outer tissues, as model systems. We are engaged in identifying small-molecule compounds that disrupt polarity, exploring their molecular targets, and using biochemical approaches to discover novel polarity-localized proteins and analyze their functions.
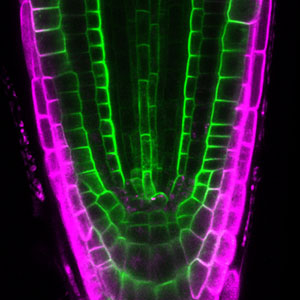
Fig. 1. Cell polarity in Arabidopsis thaliana root tip. Borate exporter BOR1-GFP (green) and boric acid channel mCherry-NIP5;1 (magenta) localize in the lateral plasma membrane domains with polarity.
In vivo biochemistry: Protein engineering and biosensor development
Understanding cellular functions in plant developmental processes and responses to the environment requires real-time quantification of intracellular biochemical and biophysical processes with high-resolution spatial information. Our research group systemically engineers biosensors utilizing fluorescent proteins to facilitate the quantitative analysis of biomolecules such as sugars and plant hormones. We are also working with a group of Chemistry at ITbM to develop imaging tools using fluorescent small molecules (Fig. 2). Moreover, we are also developing tools employing a synthetic approach to regulate plant cellular processes precisely, thereby unraveling the intricacies of metabolic signaling networks.

Fig. 2. Arabidopsis root with fluorescence dye (Cyan:Microtubule) and Fluorescent proteins (Blue:Nuclei, Magenta:Cell Membrane).
Membrane transporter–substrate–inhibitor interactions
Membrane transporters and enzymes typically exhibit broad substrate recognition rather than strict specificity. Several transporters, including SWEET sugar transporters and members of the NRT/PTR family, which are known to mediate nitrate and peptide transport, have also been shown to function in phytohormone transport (Fig. 3). Our group collaborates with other research teams at ITbM to investigate the molecular mechanisms underlying substrate selectivity in transporters with multiple substrates and to elucidate the physiological significance of each substrate. We also leverage compound libraries and advanced equipment at research centers to identify transporter inhibitors, which we are developing as novel molecular tools for application in agricultural research.
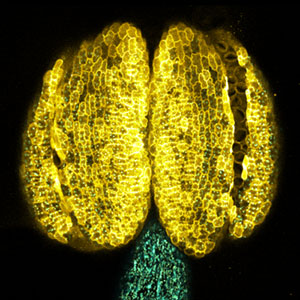
Fig. 3. Sucrose transporter SWEET13-GFP (yellow) in Arabidopsis anther. SWEET13 is required for sugar transport for pollen maturation.
References
1. Yoshinari A. et al. Plant Cell Physiol. 65, 1801 (2024)
2. Yoshinari A. et al. Plant J. 118, 1699 (2024)
3. Isoda R. et al. PNAS 119, e2207558119 (2022)
4. Yoshinari A. et al. Plant Cell 33, 420 (2021)
5. Iwatate RJ. et al. Plant Cell 32, 3081 (2020)
Member
-

Designated Lecturer
Akira Yoshinari
yoshinari.akira.t2(at)f.mail.nagoya-u.ac.jp

Group of Photobiology
Exploring the potential capacity of plants using light technology
Plants spend their entire lives where they germinate despite various environmental condition such as rain, wind, cold and heat. Throughout the long history of evolution, plants have evolved sophisticated response to adapt to changing environments. In our group, we focus on studying various environmental response in plants, such as light and mechanical stimulation. We emphasize the continuous observation of living cells and perform live-cell analysis based on cutting-edge imaging techniques employing organic fluorescent molecules and microdevice technologies. We aim to discover fascinating phenomena that captivate many people’s attention and elucidate their underlying mechanisms.
The mechanism of elongation and its deformability in plant cells
We study the response of plant cells when encountering obstacles. In nature, plant cells may encounter spaces smaller than their own size. For instance, pollens adhering to the stigma extends pollen tubes through intricate and narrow spaces within the female tissues in order to produce seeds for the next generation. In the soil, root hairs extend through the narrow gaps between soil particles to absorb water and nutrients. In both cases, the pathways for pollen tubes or toot hairs are not pre-existing. The question is how these cells behave when encountering spaces narrower than their own size. We have discovered that pollen tubes and root hairs have the ability to deform themselves and pass through gaps as small as 1 micrometer using microfabrication techniques (Fig. 1). Indeed, plant cells possess a turgor pressure comparable to that of a car tire, serving as the force for elongation. When pollen tubes and root hairs elongate within tissues or soil, it is believed that they constantly regulate the rigidity of the cell walls and turgor pressure effectively. We are conducting research to discover and unravel the mechanisms behind the potential resilience and strength of plant cells.
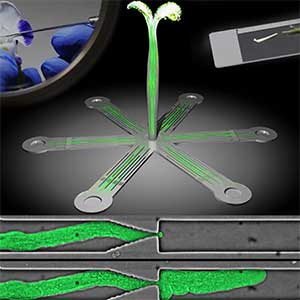
Fig. 1. Pollen tubes passing through narrow gaps within microdevices.
Mechanism of organelle positioning in plants
We are conducting research to elucidate the regulatory mechanisms controlling the number and distribution of organelles, especially in chloroplasts (Fig. 2). In cells constituting plant body, there are organelles responsible for cellular functions, and their number and distribution are properly regulated to be in the right place, at the right time, and in the right quantity. For instance, the luminous moss, which inhabits dark rock crevices, is known to arrange chloroplasts around the nucleus within specialized rounded form cells, also known as lens-shaped cells. However, the mechanism behind the formation of the lens-shaped cells and the positioning of chloroplasts remains unknown. We are focusing on the microtubule- and actin-associated proteins involved in the regulation of the number and distribution of organelles.
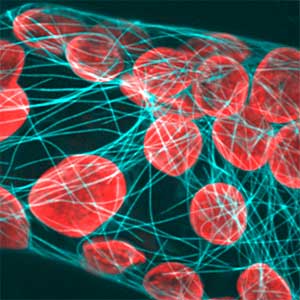
Fig. 2. Chloroplasts and microtubules in the moss Physcomitrium patens.
The mechanism of regeneration capacity in plants
We are also conducting research to unravel the mystery behind the remarkable regeneration capacity in plants. Indeed, plants exhibit a high regeneration capacity, as evidenced by the sprouting of shoots from stumps and propagating by cuttings. These abilities of plants depend on high capacity of differentiated cells to transform into pluripotent stem cells (reprogramming ability). It is known that plant hormones such as auxin and cytokinin are involved in the reprogramming of plants, but the process is still largely unknown. We are employing single-cell analysis devices, AI robotics technology, plant hormone imaging techniques, and other molecular tools to investigate the mechanisms underlying the regeneration capacity of plants.
References
1. Uno K. et al. Nat. Commun. 12, 2650, 2021
2. Yanagisawa et al. Sci. Rep. 7, 1403, 2017
3. Sato Y. et al. Sci. Rep. 7, 1909, 2017
4. Sato Y. et al. J. Biosci. Bioeng. 120, 697, 2015
5. Wada M. et al. Annu. Rev. Plant Biol. 54, 455, 2003
Member
-

Designated Associate Professor
Yoshikatsu Sato
sato.yoshikatsu.h7(at)f.mail.nagoya-u.ac.jp
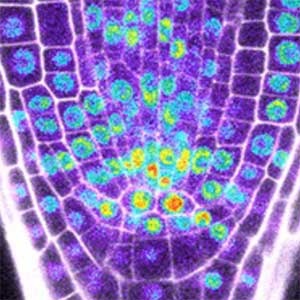
Group of Plant Pattern Formation
Unraveling the mechanistic insight of plant tissue formation
Land plants are exposed to a wide range of environmental stresses, such as temperature fluctuations, drought, and nutrient deficiencies. However, they have an exceptional adaptive capacity to cope with these challenges. Our group aims to gain a comprehensive understanding of how plants sense external stimuli and utilize this information to shape their body plan. This will contribute to our understanding of the mechanistic insights into plant survival strategies, including how plants sense and adapt to environmental changes, and will provide important implications for the fields of plant ecology, agriculture, and environmental protection.
Understanding how peptide molecules act as messengers in plant responses to environmental stresses
Plants have a remarkable ability to communicate with the signals from their surroundings, and effectively employ these signals to protect themselves and to control their own developmental strategies. Our study delves into the intricate processes through which plants detect environmental changes using a range of signaling pathways, leading to various physiological reactions. Through this exploration, we aim to reveal fresh perspectives on how plants ensure their survival, which are instrumental in increasing crop productivity and their resilience to changing environmental dynamics.
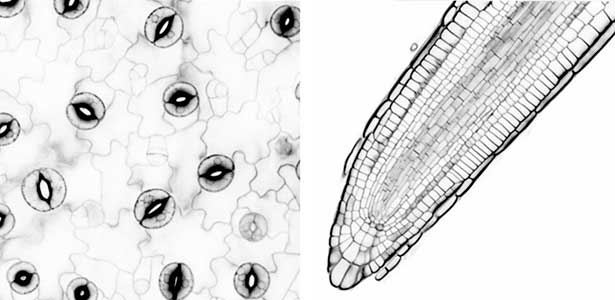
Fig. 1. Leaf epidermal tissue (left) and root apical meristem (right) of Arabidopsis thaliana.
Understanding the molecular mechanisms governing plant stem cell activation in response to environmental cues
Plant stem cells play a critical role in the growth and vitality of plants, especially in adapting to environmental changes. These cells exhibit an extraordinary capacity to regenerate new tissues. Precise regulation of stem cell activities is essential to maintain and optimize plant development. Our current research focuses on identifying key regulators crucial for initiating and maintaining plant stem cell activities. Furthermore, we aim to investigate the intricate interactions of these factors and how they are influenced by environmental stimuli. This study seeks to not only elucidate the fundamental mechanisms governing plant stem cell regulation but also offers potential for the creation of novel plant varieties with improved environmental resilience.
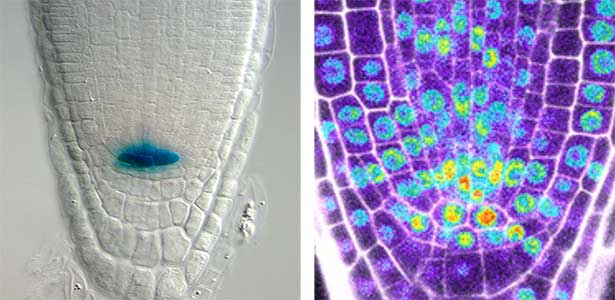
Fig. 2. WOX5 expression in the quiescent centre (left) and false color representation of gene expression patterns related to root stem cell activity (right).
References
1. Shimotohno A. Plant Biotech. 39, 19-28, 2022.
2. Shimotohno A. et al. Annu. Rev. Plant Biol., 72, 273-296, 2021.
3. Shimotohno A. and Scheres B. Curr. Opin. Plant Biol. 51, 74-80, 2019.
4. Shimotohno A. et al., Genes & Dev. 32, 1085-1100, 2018.
Member
-

Designated Lecturer
Akie Shimotohno
ashimo(at)itbm.nagoya-u.ac.jp
G30 International Program
Group of Microbial Motility
Explore the mechanism of motility using bacteria
Microorganisms are much simpler creatures than higher eukaryotes like plants and animals, including humans. But microorganisms are equipped with minimal mechanisms essential for life, and have survived ever since the moment of birth of life on the earth. Evolution allowed higher eukaryotes to develop mechanisms to achieve their complex life styles/functions, but such complexity is based on the simpler ones already existing in microorganisms. In other words, the basic principles in life can be clarified by studying the mechanisms of microbial systems. In our laboratory, we are trying to elucidate the mechanism of "motility", one of fundamental phenomena of life, using unicellular bacteria with flagella as the motility organ.
Bacterial motility organ: flagellum
Cell motility is a fundamental activity of life, and the elucidation of the operating principles of the apparatus responsible for motility is a quite important question of life science. Bacteria, which consist of only one cell, use motility apparatus as a means of escaping from toxic substances and moving toward nutrient in order to survive in the environment. Many bacteria use the flagellum as motility apparatus, with spiral flagellar filaments rotated by a motor embedded in the cell surface layer at its base to generate the driving force for motility.
Mechanism of energy conversion and rotation in bacterial flagellar motors
The flagellar motor (Fig. 1) harnesses electrochemical gradient of ions across the cell membrane (ion driving force) to generate rotational force. The flagellar motor is ~ 50 nm size of supramolecular membrane complex. What makes unique about this motor is that it can convert, not ATP, but transmembrane ion motive force to rotational force. Such an ion-driven rotary motor does not exist in eukaryotic organisms. Furthermore, this motor is a high-performance molecular machine that can rotate as fast as 1000 revolutions per second in both directions, change direction instantly, and adjust its energy conversion capacity by sensing the load. Although the flagellar motor has attracted attention across the fields of science, engineering, and medicine, much of the energy conversion mechanism is still in a mystery. We are attempting to clarify the rotational mechanism by focusing on both the stator, which is the energy conversion unit, and the rotor ring, which is involved in controlling the direction of rotation, and elucidating the relationship between structure and function.
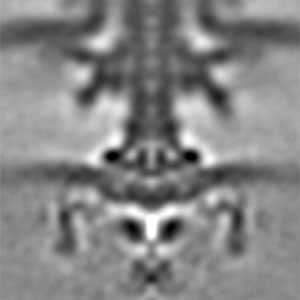
Fig. 1. The cryo-electron tomographic image of the flagellar motor of marine Vibrio.
Regulation of the number and location of flagellar formation
Our bodies are shaped by functional molecules which are under appropriate spatiotemporal control during the process of development. In addition, in order to maintain tissues and organs normally, those molecules must work in appropriate amounts at each site. If this mechanism fails, it may cause disease. Thus, placement of biomolecules in appropriate amounts at functional sites is a fundamental function of all living organisms. In bacteria, the number and position of flagella are strictly determined to function optimally, so that each cell can move appropriately in the environment in its habitat. Using marine Vibrio as a model organism, which form a single flagellum at the pole of the cell, we are trying to find out why “only one” flagellum can be formed at the special position called the “pole of the cell” (Fig. 2). Instead of using complex living organisms, we use simple and fast-growing bacteria to conduct research aimed at elucidating the mechanism of the proper arrangement of biological supramolecules.
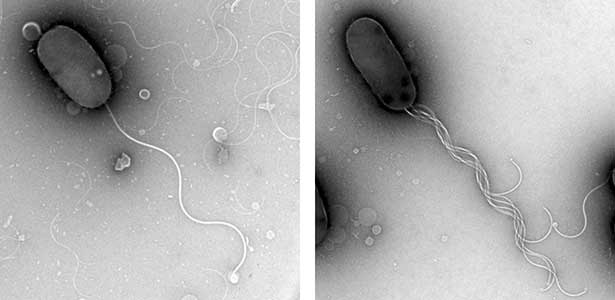
Fig. 2. The electron micrographic image of wild-type Vibrio cell with only one polar flagellum at cell pole (left) and a mutant with multi-polar flagella at a cell pole (right).
References
1. Nishikino T. et al. PNAS 122, e2415713122 (2025)
2. Takekawa N. et al. mBio 15, e0126124 (2024)
3. Kojima S. et al. Structure 26, 590 (2018)
4. Ono H. et al. Mol. Microbiol. 98, 130 (2015)
5. Zhu S. et al. PNAS 111, 13523 (2014)
Member
-

Professor
Seiji Kojima
z47616a(at)cc.nagoya-u.ac.jp
Molecular and neuronal mechanism of behavioral aging
Our neuronal functions decline with aging. Accumulation of DNA damage, oxidative stress, and diseases such as Alzheimer's disease have been proposed as mechanisms of age-dependent decline of neuronal functions. However, can these passive mechanisms explain the differences in aging speed among species? We hypothesize that the genetic program may actively cause neuronal aging and have been attempting to elucidate the mechanism using the nematode Caenorhabditis elegans (C. elegans) as a model.
Molecular mechanisms of behavioral aging
The major challenges to studying the aging of neuronal functions are the long aging time and the complexity of the nervous system. Therefore, we use the nematode C. elegans, which has a short lifespan and a simple nervous system, as a model. C. elegans becomes an adult within three days after fertilization, undergoes a reproductive period of a few days, and completes its ephemeral life span in about two weeks (Fig. 1). During this time, it exhibits various behaviors using only 302 neurons. Many of these behaviors diminish around the time they finish reproducing. The existence of a genetic program for aging is suspected to cause behavioral changes because age-dependent behavioral decline happens in only one week in C. elegans, whereas in humans, it spans several decades. We, therefore, conduct novel genetic screening for mutants that do not exhibit age-dependent behavioral decline and analyze these mutants. Since molecular mechanisms of critical biological processes are often shared between C. elegans and mammals, we hope our research will elucidate the fundamental principles of neuronal aging.
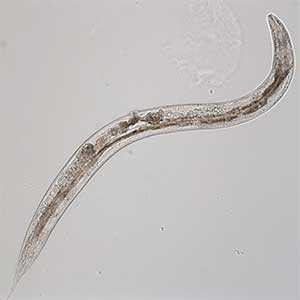
Fig. 1. Aged C. elegans (10-day-old adult).
Effect of diet on behavioral aging
Not only genetic programs but also environmental factors such as diet affect the aging of neuronal function. We found that bacteria, the food of C. elegans (Fig. 2, left), influence behavioral aging. To elucidate the neural basis of these diet- and aging-induced behavioral changes, we measure neuronal activity manifested as changes in intracellular calcium concentration. Because C. elegans is translucent (Fig. 2, right), we can readily monitor the neuronal activity of a live animal using a protein whose fluorescence intensity reflects calcium concentration.

Fig. 2. Bacteria in the C. elegans intestine (green, left) and neuronal nuclei in the C. elegans head (red, right).
Biological significance of behavioral aging
The behavioral decline in C. elegans is mainly observed in behaviors that navigate animals toward bacterial food. Perhaps the alteration of neural function due to aging and the accompanying behavioral changes may be a mechanism by which aged individuals do not compete with younger individuals for food, maximizing the group’s benefits at the individual’s expense. C. elegans is as tiny as one mm long, making it a powerful tool for population analyses. In the future, we would like to explore the significance of aging in populations using mutants less prone to behavioral aging obtained through our genetic screening.
References
1. Aleogo BM. et al. PNAS 122, e2412391122 (2025)
2. Suryawinata N. et al. Sci Rep. 14, 5529 (2024)
3. Higurashi S. et al. eLife 12, e81418 (2023)
4. Noma K. et al. eLife 6, e26376 (2017)
5. Noma K. and Jin Y. Nature Commun. 6, 8868 (2015)
Member
-

Associate professor
Kentaro Noma
noma.kentaro.f1(at)f.mail.nagoya-u.ac.jp
Graduate School of Pharmaceutical Sciences
Group of Structural and Molecular Pharmacology
Controlling three-dimensional structure for new drug discovery
Medicines are a fundamental item of medical care and protection of our health. In addition, medicines also contribute to maintain our economic and cultural activities from the threat of disease. However, there are still many patients are waiting for the cure for their disease to be perfected. In many disease cases, the current standard of care entails a significant burden on the patient. In order to satisfy such "unmet medical needs", the scientists must establish new drug discovery methods with a cutting-edge technology, that utilize three-dimensional structural information of molecules. Professor Hiroaki's group is engaging in a new drug discovery method utilizing protein structure and NMR information, and Associate Professor Kodama's group is researching the design and synthesis of novel artificial nucleic acid molecules and their practical application for therapeutics.
Discovery and development of a novel anticancer drug candidate NPL-3009
Triple negative breast cancer (TNBC) is a type of breast cancer that is resistant to common hormone therapy and molecular targeted drugs (Herceptin). TNBC accounts for 15 to 20% of breast cancers with poor prognosis. It has been reported that the Wnt signaling pathway is over-activated in TNBC, so that we have started to search for inhibitors of Dvl that may block Wnt signaling, using the molecular structure of Dvl as a clue. During this project, we succeeded in elucidating the structure of the new inhibitor named NPL-3009 bound to the Dvl-PDZ. This achievement is expected to contribute to the design and development of new anti-cancer drug molecules (Fig. 1).

Fig. 1. Two conformations in the crystal of the anticancer drug candidate, “NPL-3009” (left) and the complex structure with the Dvl1-PDZ domain (right).
Nucleic acid drug candidate molecules with a new skeleton
The Nobel Prize in Physiology or Medicine 2023 was awarded to Drs. Catalin Carrico and Drew Weissman, for their “discovery of nucleic acid modifications that enabled the development of an effective mRNA vaccine against COVID-19”. Natural RNA and DNA molecules are known to be easily and rapidly degraded in vivo, making it difficult for them as pharmacologically active therapeutics. We have designed and synthesized many nucleosides with the designed sugar moieties. These modified nucleosides will be incorporated into oligonucleic acid molecules, and further analyzed their conformation when bound to the target mRNA.
New cytoskeleton formed by human red blood cell proteins
Stomatin (Band 7.2 protein) is a membrane protein that is highly abundant in erythrocyte membranes. The SPFH domain is a domain of unknown function in the central part of the stomatin. We eventually discovered that the domain can form linear fibrils (Fig. 2). It is possible that changes in physiological conditions cause stomatin to assemble and act as a cytoskeleton just beneath the erythrocyte membrane to reguralte membrane stiffness.
Fig. 2. Electron microscopic image of fibers formed by the SPFH domain of the human erythrocyte protein Stomatin.
References
1. Miura K. et al. J. Virol. 13, e0042623 (2023)
2. Emoto Y. et al. Methods Protoc. 6, 44 (2023)
3. Mizoguchi T. et al. Int. J. Mol. Sci. 24, 4313 (2023)
4. Kataoka K. et al. Curr. Res. Struct. Biol. 4, 158 (2022)
5. Ikeda K. et al. Sci. Rep. 10, 12334 (2020)
6. Moai Y. et al. Nucleosides Nucleotides Nucleic Acids 39, 131 (2020)
Member
-

Professor
Hidekazu Hiroaki
hiroaki.hidekazu.j7(at)f.mail.nagoya-u.ac.jp
Structural biology of protein-drug interactions
-

Associate Professor
Tetsuya Kodama
kodama(at)ps.nagoya-u.ac.jp
Creation of novel nucleic acid therapeutics with controlled steric structure
-

Assistant Professor
Emi Hibino
hibino.emi.r3(at)f.mail.nagoya-u.ac.jp
Research on protein aggregation mechanisms and search for aggregation inhibitors
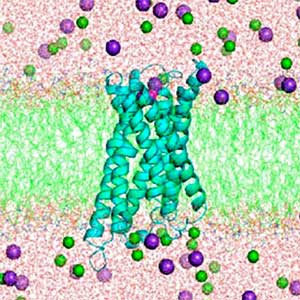
Group of Cellular and Structural Physiology
Study of the signal transduction mechanism across biomembranes
Membrane proteins such as channels, pumps, and G protein-coupled receptors are associated with various biological events and homeostasis through their roles in neurotransmission, intercellular communication, and ion transport across the biomembranes. High-resolution structural analysis and functional studies of these membrane proteins lead to our understanding of the molecular basis of biological phenomena in vivo. We provide education and research based on structural biology, biochemistry, cell biology, and computer science.
Structural studies and gating mechanism of cel-cell junction channels
Gap junction channels are known to be direct communication channels between adjacent cells, but the structural basis for the gating mechanisms remains unclear. We have reported the cryo-EM structures of the Caenorhabditis elegans innexin-6 gap junction channels. By comparing the structures of the solubilized and lipid nanodisc-reconstituted states, we have proposed a gating model in which lipid diffusion occludes the channel passage (Fig. 1). Multiple approaches, including structural biology, functional analysis, and computational science, will help to gain a deeper understanding of signal transmission across biomolecular membranes.
Fig. 1. Cryo-EM image and 3D reconstruction of a gap junction channel.
The functional analysis of channel molecules in morphogenesis and pattern formation
It has become increasingly clear that channel molecules present in the cell membrane play a role in animal body shape formation. In particular, gap junctions, which facilitate the direct exchange of small molecules and ions between adjacent cells, contribute to pattern formation, such as the repetitive structures seen in animal skin patterns and vertebral bones (Fig. 2). This study aims to analyze how the electrical properties of channels influence animal morphogenesis.

Fig. 2. Gap junction channels associated with stripe pattern formation.
Computational design and discovery of novel proteins
Computers can simulate nature in model-based manners and process massive data beyond human ability. The latter allows us to step into the regions in vast state spaces, such as that of possible amino acid sequences, already sampled by natural evolution but uncharted by humans; this ability can be redirected to discover naturally occurring proteins unseen by human beings. The former ability to mimic natural phenomena, like protein folding, allows us to explore the unexplored even by nature; this enables us to design novel artificial proteins whose natural counterparts have never emerged in evolution. Exploration of these two classes of the unknown would contribute to expanding human knowledge.
References
1. Sakuma K. et al. Protein Sci. 33, e4934 (2024)
2. Suzuki S. et al. Commun. Biol. 5, 707 (2022)
3. Usui Y. and Watanabe M. BBA-Adv. 1, 100006 (2021)
4. Burendei B. et al. Sci. Adv. 6, eaax3157 (2020)
5. Oshima A. et al. Nature Commun. 7, 13681 (2016)
Member
-

Professor
Atsunori Oshima
atsu(at)cespi.nagoya-u.ac.jp
Gating mechanism of channel proteins responsible for intercellular communications
-

Associate Professor
Masakatsu Watanabe
watanabe.masakatsu.j2(at)f.mail.nagoya-u.ac.jp
The functional analysis of channel molecules in morphogenesis and pattern formation
-

Assistant Professor
Koya Sakuma
sakuma.koya.u6(at)f.mail.nagoya-u.ac.jp
Computational design and discovery of novel proteins
