RESEARCH
Tracing back from molecules to mechanisms
Peptide hormones and receptors in plants
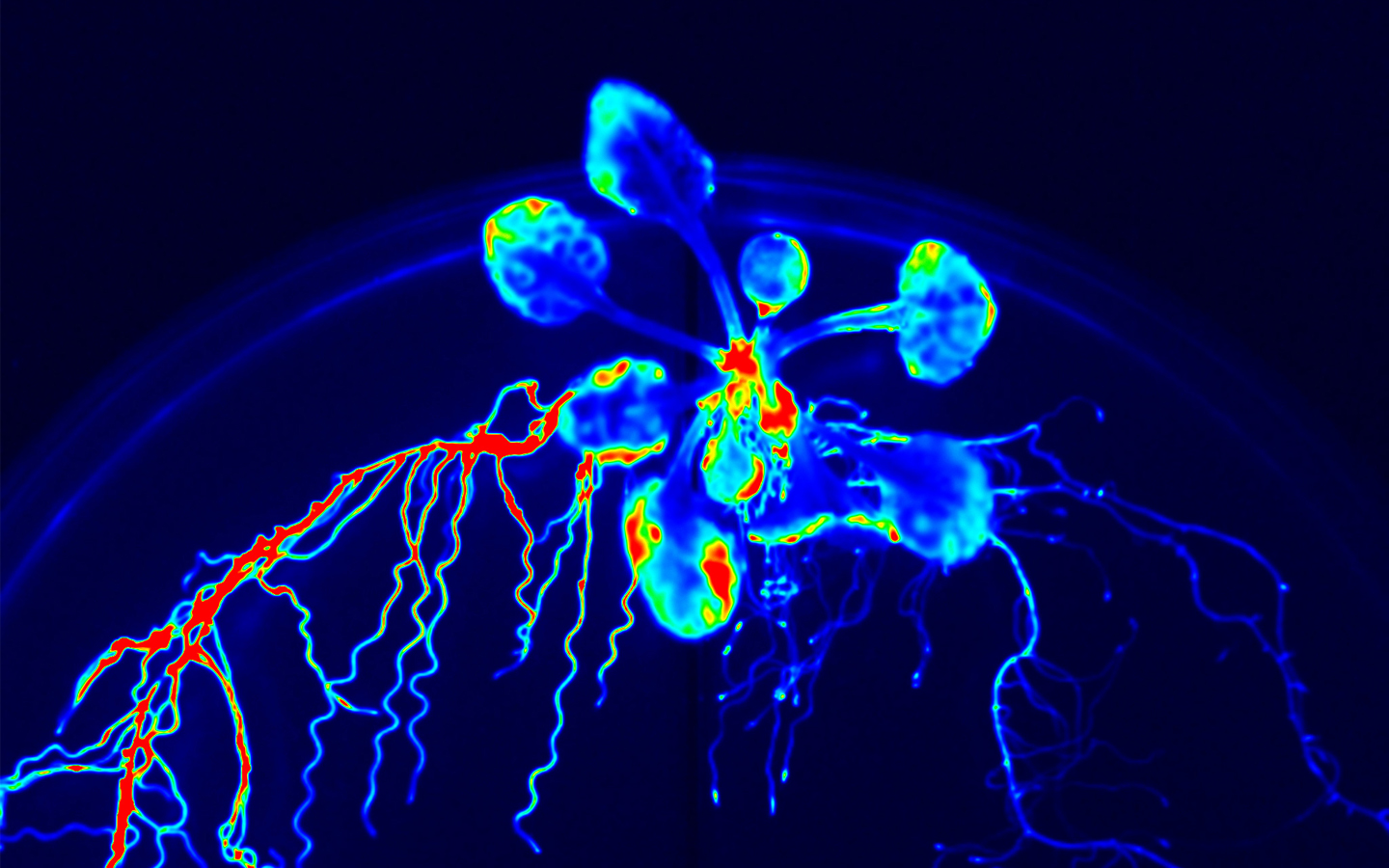
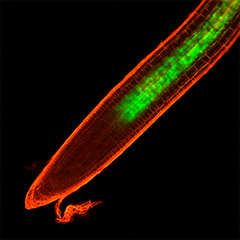
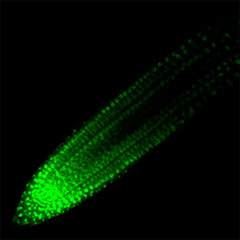
In biology, the general methodology is to first focus on a specific biological phenomenon, then to identify the key molecules involved, and finally to explain the mechanism. However, if we first focus on a specific molecule, analyze its function, and finally aim to discover new mechanisms and phenomena, a new biology may be born. We have been searching for novel peptide hormone-receptor pairs by using the following strategies: (1) in silico identification of genes encoding small secreted peptide hormones, (2) determination of their mature peptide structures by mass spectrometry, and (3) identification of their receptors by binding experiments using an expression library of receptor candidates. This approach has led to the discovery of peptide hormones involved in growth and environmental responses, including RGF (Science 2010), which regulates activity and patterning of root apical meristem; CEP (Science 2014), which mediates systemic regulation of nitrogen acquisition; CIF (Science 2017), which is involved in Casparian strip development (Science 2017), and PSY (Science 2022), which regulates the trade-off between growth and stress responses.
Post-translational modifications
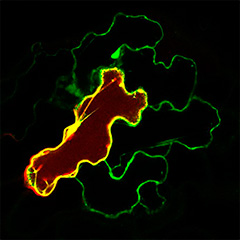
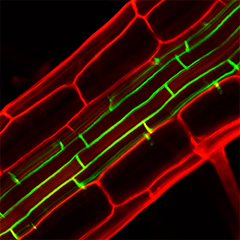
Post-translational modifications of peptides also provide important clues for tracing back from molecules to mechanisms. Because post-translational modifications generally have a critical effect on peptide function, phenotypic analysis of a mutant deficient in a post-translational modification enzyme provides an overall picture of the function of a group of post-translationally modified peptides. For example, mutants deficient in tyrosine sulfotransferase have shorter roots and dwarf shoots (PNAS 2009), a phenotype that was a major clue to the discovery of peptide hormones such as RGF and PSY. In mutants deficient in enzymes involved in extracellular protein glycosylation such as Hyp O-arabinosylation (Nature Chem. Biol. 2013) and Hyp O-galactosylation (Plant J. 2015), various tissue defects have also been observed, suggesting the existence of as-yet-unknown extracellular signaling mechanisms. In particular, Hyp O-galactosylation is an essential modification of arabinogalactan protein (AGP) function. Mutants deficient in this modification are unable to generate functional AGPs, which allows us to overcome the gene redundancy problem and gain a better understanding of the functions of AGPs.
Phosphorylation/dephosphorylation-mediated signaling
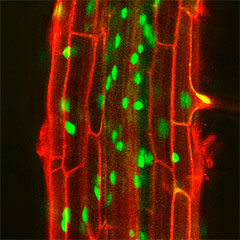
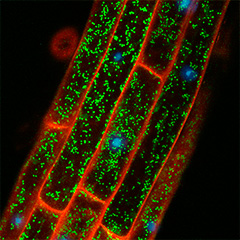
When a peptide hormone binds to a receptor on the cell membrane, the intracellular signal transduction system through protein phosphorylation and dephosphorylation is activated. Tracking this phosphorylation and dephosphorylation process tells us how cell signaling works. In addition, there are still many protein phosphatases and kinases in plants whose functions remain unknown, and the search for their substrates is also an attractive research target that will lead to the elucidation of new mechanisms. Using our improved 15N metabolic labeled quantitative phosphoproteomics system, we can identify the substrates of the protein phosphatases. For example, PP2C family phosphatase, CEPH, dephosphorylates the Ser501 residue of NRT2.1, a major nitrate transporter in roots, which turns on nitrate uptake activity (Nature Plants 2021).
Phloem-mobile non-secreted peptides
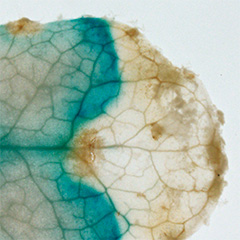
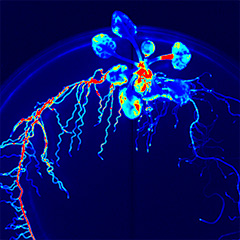
Our recent studies have revealed that non-secreted peptides such as CEPD (Nature Plants 2017) and CEPDL2 (Nature Commun. 2020), which are induced in leaves in a nitrogen deficiency-dependent manner, translocate to roots through phloem and regulate the expression of nitrate uptake transporters. Phloem is a vascular tissue comprised of sieve-tube element cells, with numerous pores, called sieve pores, at the cell-cell interface, allowing the cytoplasm inside to migrate between cells. Therefore, non-secreted peptides in the phloem can migrate long distances between organs. This is a mechanism unique to plants that is not found in animals, and there are still many unknowns. Analysis of a group of non-secreted peptides that exhibit similar phloem-mobile nature may reveal dynamic communication between plant organs.