II. DEVELOPMENT
OF ARABIDOPSIS LEAVES:
PRINCIPLE BEHIND LEAF
SYMMETRY
Recent
references
Semiarti
et al., Development (2001)
Iwakawa
et al., Plant & Cell Physiol. (2002)
The asymmetric leaves2 (as2) mutant of Arabidopsis
thaliana generated leaf lobes and leaflet-like structures from the
petioles of leaves in a bilaterally asymmetric manner. Both the delayed
formation of the primary vein and the asymmetric formation of secondary
veins were apparent in leaf primordia of as2 plants. A distinct
midvein, which is the thickest vein and is located in the longitudinal
center of the leaf lamina of wild-type plants, was often rudimentary even
in mature as2 leaves. However, several parallel veins of very
similar thickness were evident in such leaves. The complexity of
venation patterns in all leaf-like organs of as2 plants was reduced.
The malformed veins were visible prior to the development of asymmetry
of the leaf lamina and were maintained in mature as2 leaves.
Culture in vitro on phytohormone-free medium of leaf sections from as2
mutants and from the asymmetric leaves1 (as1) mutant, which
has a phenotype similar to that of as2, revealed an elevated potential
in both cases for regeneration of shoots from leaf cells. Analysis
by the reverse transcription-polymerase chain reaction showed that transcripts
of the KNAT1, KNAT2 and KNAT6 (a recently identified
member of the class 1 knox family) genes accumulated in the leaves
of both as2 and as1 plants but not in wild-type leaves.
Transcripts of the STM gene also accumulated in as1 leaves.
These findings suggest that, in leaves, the AS2 and AS1 genes
repress the expression of these homeobox genes, which are thought to maintain
the indeterminate cell state in the shoot apical meristem. We propose
that AS2 and AS1 might be involved in establishment of a
prominent midvein and of networks of other veins as well as in the formation
of the symmetric leaf lamina, which might be related to repression of expression
of class 1 knox homeobox genes in leaves.
We have characterized the AS2 gene, which
appears to encode a novel protein with cysteine repeats (designated the
C-motif) and a leucine-zipper-like sequence in the amino-terminal half
of the primary sequence. The Arabidopsis genome contains 42
putative genes that potentially encode proteins with conserved amino acid
sequences that include the C-motif and the leucine-zipper-like sequence
in the amino-terminal half. Thus, the AS2 protein belongs to a novel
family of proteins that we have designated the AS2 family. Members
of this family except AS2 also have been designated ASLs (AS2-like proteins).
Transcripts of AS2 were detected mainly in adaxial domains of cotyledonary
primordia. Green fluorescent protein-fused AS2 was concentrated in
plant cell nuclei. Overexpression of AS2 cDNA in transgenic
Arabidopsis plants resulted in upwardly curled leaves, which differed
markedly from the downwardly curled leaves generated by loss-of-function
mutation of AS2. Our results suggest that AS2 functions in
the transcription of a certain gene(s) in plant nuclei and thereby controls
the formation of a symmetric flat leaf lamina and the establishment of
a prominent midvein and other patterns of venation.
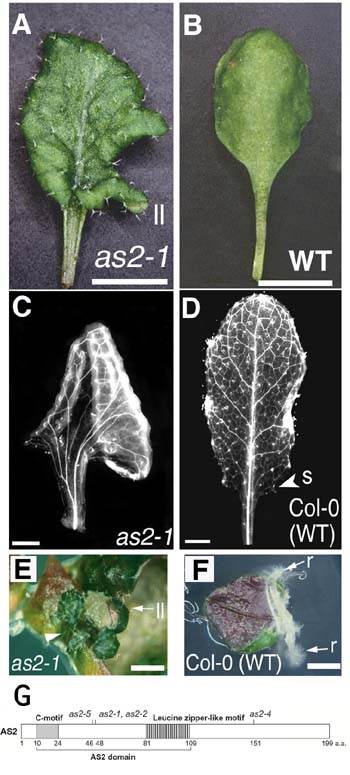
|
Figure
(A - D) Characteristic features of leaves of the asymmetric leaves2
(as2) mutant. A, leaf shape of as2; B, leaf shape of
wild type; C, venation pattern of as2; D, venation pattern of wild
type. (E and F) The generation of shoots on as2 leaves in
vitro. Sections of the as2 (E) and wild-type (F) leaves were
photographed after incubation for 16 days on hormone-free MS medium.
Several percent of leaf sections from as2 have produced shoots at
the sinus of a leaf lobe while no section from the wild-type leaf have
generated shoots but roots at the efficiency of 12% (see Semiarti et al.,
2001). (G) Domain organization and characteristic features of the
predicted AS2 protein. The region indicated by a bracket below the
box is the AS2 domain. The shaded box and the striped box represent
the C-motif and the leucine-zipper like sequence, respectively. Sites
of as2-1, as2-2, as2-4, and as2-5mutations are indicated.
Numbers below the box indicate positions of amino acid residues. |
|
